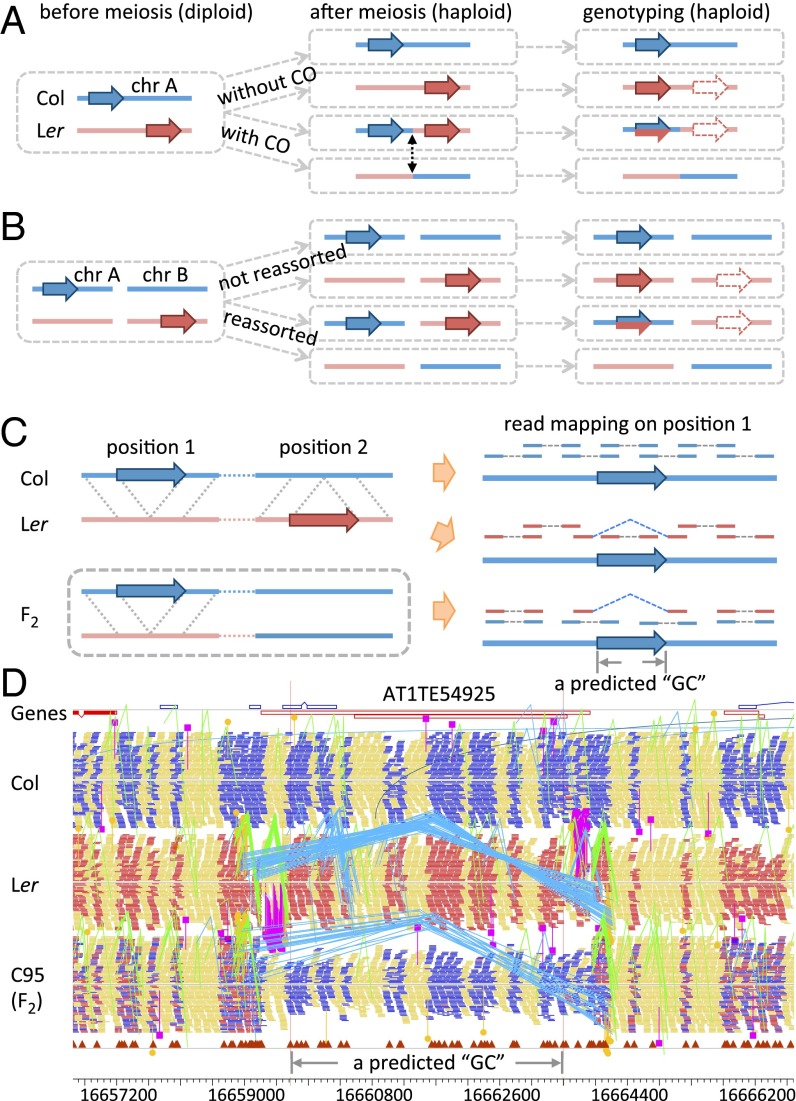
Fig. 2.
Paralogous sequences between two ecotypes and their effects on allele ratio estimation. Redistribution of SV-related paralogous DNA segments in meiotic progenies and consequent genotyping using short read mapping for paralogs on the same chromosome (A), or paralogs on different chromsomes (B). Col and Ler are marked in the same colors as in Fig. 1, whereas dashed arrows indicate actual sites of short reads which were misplaced at nonallelic sites in the reference genome (Col). (C) Read depth, mapping distance, and orientation of PE reads from Col, Ler, and the F2 on the reference genome (Col) around the SV (large blue arrows in both Left and Right). On the Right, PE reads are shown above the Col (blue) sequences; Col read pairs and most Ler read pairs are mapped normally, except those Ler read pairs flanking a deletion, shown here as distantly mapped red reads with blue linkers. Only one chromatid is shown for both Col and Ler. An F2 plant has a heterozygous (Col/Ler) genotype around position 1 and homozygous (Col/Col) around position 2. (D) PE read mapping from Col, Ler, and an F2 plant by Yang et al. (32) in a 9-kb window (16,657,200 ∼16,666,200) on chromosome 1 of Col reference genome. Col reads mapped normally; PE mapping patterns for Ler reads indicate a deletion: A TE is surrounded by a group of reads pairs (linked by blue lines) that mapped farther apart than expected, and another two group of reads pairs (marked by pink lines) mapped to different chromosomes.