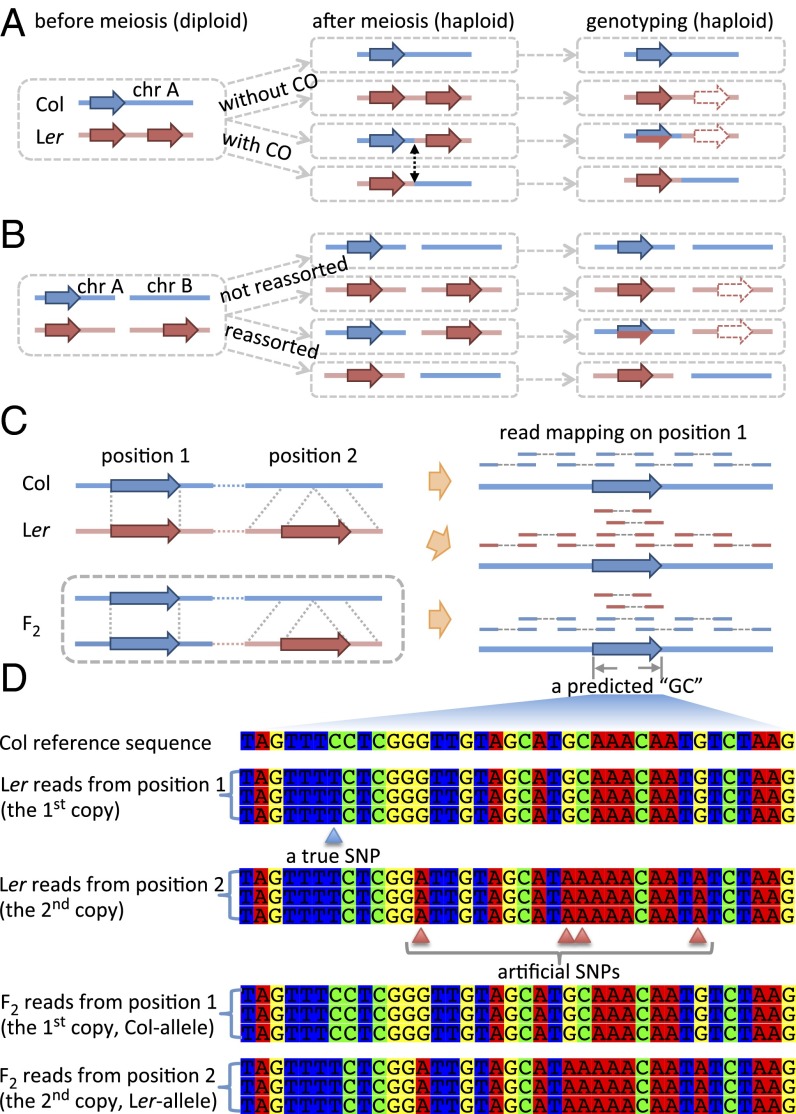
Fig. 3.
PE reads mapping patterns for CNVs and corresponding genotype predictions. Genotyping of SV-related genomic regions in meiotic progenies due to misplacement of short read in the case of CNVs on the same chromosome (A), or CNVs on different chromosomes (B). (C) Mapping pattern of reads from Col, Ler, and the F2 plant on the reference genome. The F2 plant (an example) has two Col-homologous copies and one Ler-homologous copy. Both Ler and the F2 show abnormally higher read depth within the duplicated segment. (D) Both Ler and the F2 could have two or three kinds of “genotypes,” depending on the sequence divergence of the three segments in C, where Col has one copy and Ler has two copies. Comparison of allelic sequences between Col and Ler give one true SNP (in blue), whereas misplacing the nonallelic copy of Ler creates three artifactual SNPs (in red).