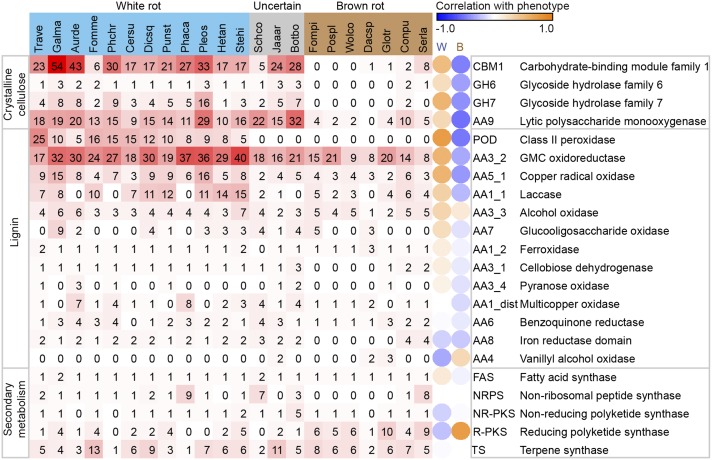
Fig. 1.
Lignocellulose-degrading and secondary metabolism in wood-decaying fungi. Organisms use the following abbreviations: Aurde, Auricularia delicata; Botbo, Botryobasidium botryosum; Cersu, Ceriporiopsis subvermispora; Conpu, Coniophora puteana; Dacsp, Dacryopinax sp.; Dicsq, Dichomitus squalens; Fomme, Fomitiporia mediterranea; Fompi, Fomitopsis pinicola; Galma, Galerina marginata; Glotr, Gloeophyllum trabeum; Hetan, Heterobasidion annosum; Jaaar, Jaapia argillacea; Phaca, Phanerochaete carnosa; Phchr, Phanerochaete chrysosporium; Pleos, Pleurotus ostreatus; Pospl, Postia placenta; Punst, Punctularia strigosozonata; Schco, Schizophyllum commune; Serla, Serpula lacrymans; Stehi, Stereum hirsutum; Trave, Trametes versicolor; Wolco, Wolfiporia cocos. Gene number is shaded red/white, and independent-contrasts correlation of enzyme with rot type is shaded orange/blue. Notice a strict white/brown-rot dichotomy with respect to the lignin-attacking PODs and the CAZymes that target crystalline cellulose (CBM1, GH6, and GH7), and a continuum with other lignin-targeting enzymes.