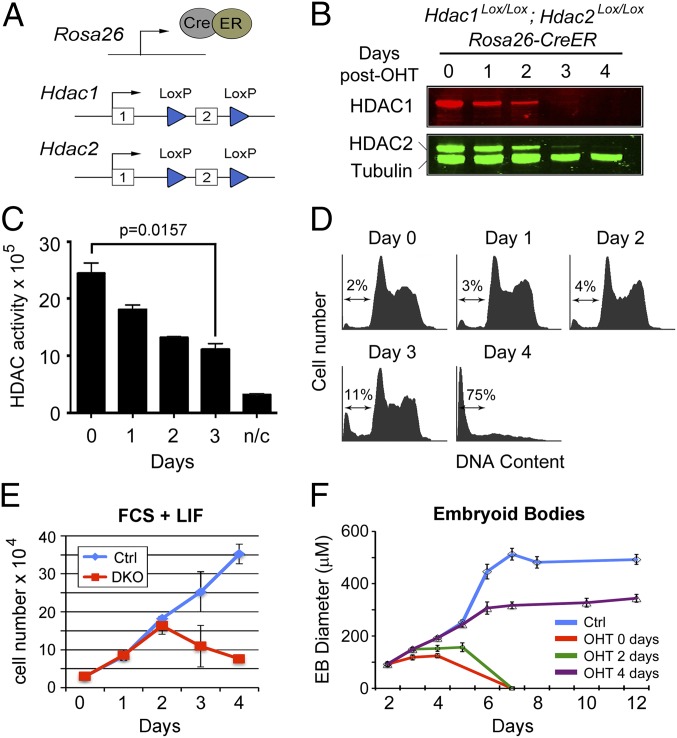
Fig. 1.
Inactivation of Hdac1/2 causes loss of cell viability. (A) Schematic diagram of the model system. An E14 ES cell line constitutively expressing a Cre/estrogen receptor (CreER) fusion from the ROSA26 locus was used to generate homozygous conditional knockout alleles for both Hdac1 and Hdac2. Both genes are inactivated by deletion of exon 2, which is flanked by LoxP sites (blue triangles). (B) Quantitative Western blot showing loss of HDAC1 and HDAC2 proteins following gene inactivation (0–4 d). Cells were cultured in the presence of 4-hydroxytamoxifen (OHT) for 24 h to induce the deletion of Hdac1/2. α-Tubulin was used to normalize protein loading, and blots were visualized and quantified using an Odyssey scanner. (C) Deacetylase activity was measured in whole-cell extracts on 4 consecutive days following OHT treatment; n/c represents the negative control. Data are representative of n > 3 independent experiments. Significance (P value) was calculated using a two-tailed t test. (D) Cell cycle distribution of Hdac1/2-deleted ES cells over a 4-d period following gene inactivation was performed using propidium iodide staining and FACS analysis. The arrow indicates the percentage of cells with a sub-G1 amount of DNA. (E) Comparative viable cell counts between control ES cells (Ctrl, untreated DKO cells) and Hdac1/2 double-knockout ES cells (DKO). All values are means (n = 3) ± SEM. (F) Embryoid bodies (EBs) were generated by plating ES cells on to bacterial dishes. OHT was add to the growth media at the indicated times following plating. The mean size of EBs size is shown (n > 30) ± SEM.