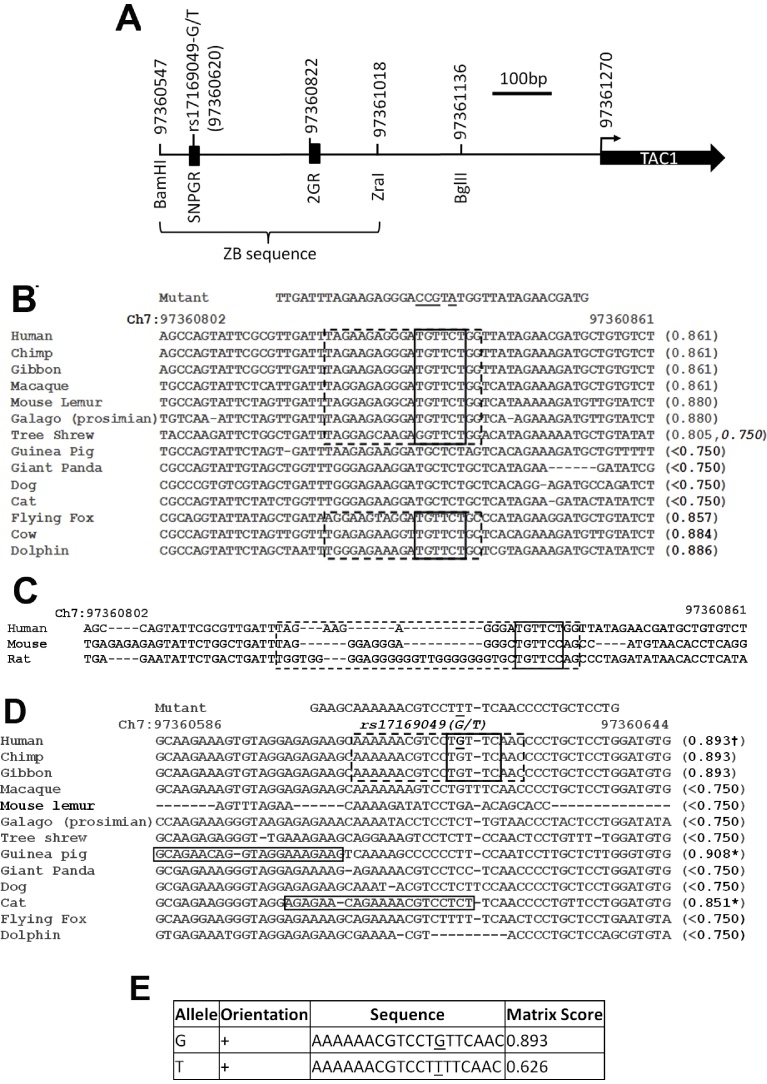
Figure 2.
(A) Scale line diagram showing features described in the text including restriction sites, putative GR binding sites (filled black boxes), the rs17169049 SNP and the TAC1 transcriptional start site (bent arrow). Numbering refers to the location of each feature on chromosome 7 based on the Human Feb. 2009 (GRCh37/hg19) assembly. (B–D) Multiple genome alignments of sequences within the TAC1 promoter that contain predicted GR binding sites (B; 2GR and C; SNPGR, binding matrices >0.75 match to Transfac GR consensii) demonstrating alignment between 13 placental mammals. Broken black boxes represent the extent of predicted GR binding matrices and solid black boxes represent matches to known GR core binding motifs. Numbers at the end of each sequence in brackets represent the degree of match to known Transfac GR binding matrices for each species. Numbers with asterisks represent the predicted match of non-homologous GR binding site matrices within the guinea pig and cat TAC1 promoters. †The binding matrix match of the T-allele of rs17169049 is reduced to 0.626. (C) Linear alignment of the human TAC1 promoter sequence around 2GR (broken black box) with the more diverged rat and mouse sequences highlighting the presence of a recognisable GR binding site (Transfac GRQ6 consensus in bold with core sequence highlighted with a solid black box). (B–D) Numbers above the alignments represent the genomic co-ordinates of human sequence (GRCh37/hg19 assembly) and numbers in D in italic represents the position of SNP rs17169049(G/T; underlined in D). Sequences labelled “Mutant” represent the sequence of oligonucleotide primers (changes underlined) used for site directed mutagenesis to produce the mut2GR (B) and the SNPGR-T variant of rs17169049 (D) as well as for the EMSA studies. (E) Table showing readout from RegSNP demonstrating predicted differences (Matrix Score) in GR binding between the G and T alleles of rs17169049.