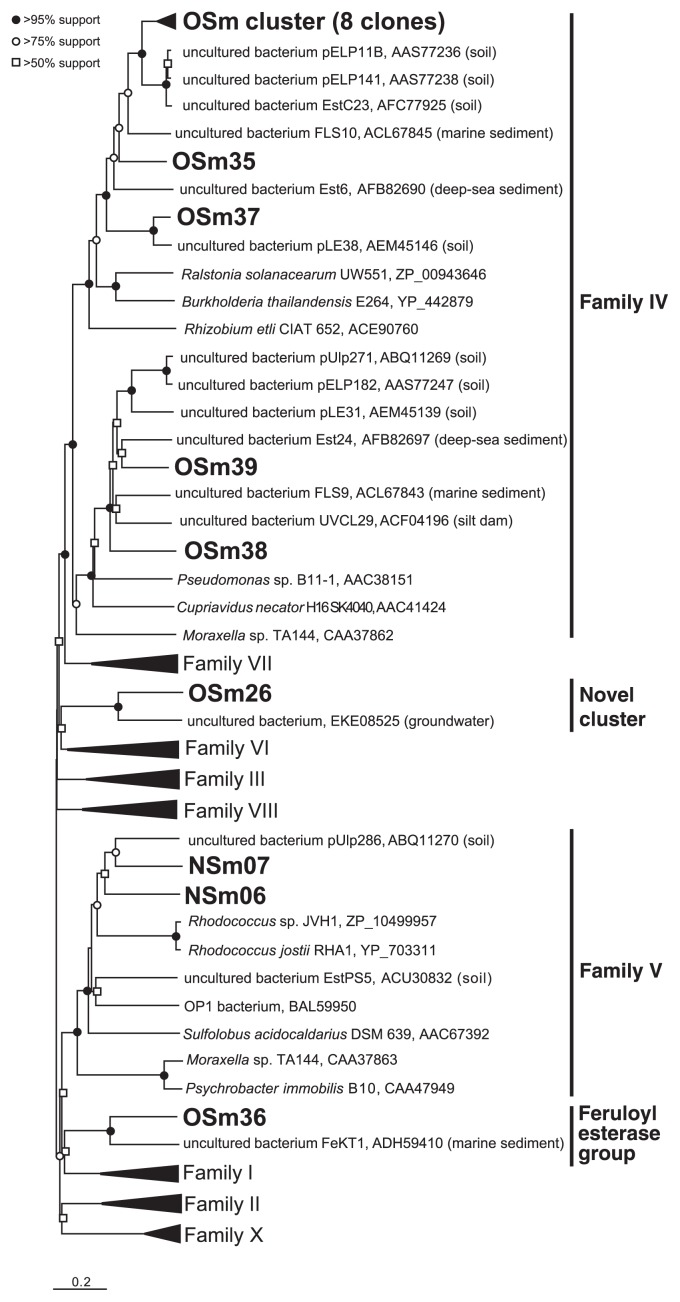
Fig. 1.
Unrooted distance matrix tree of lipolytic enzymes obtained from non-enriched soil (NS) and oil-enriched soil (OS) metagenomic libraries. The phylogenetic tree was generated by the neighbor-joining method with MEGA 5.0 software. The classification of previously known lipolytic enzymes was based on Arpigny and Jaeger (2). Eight clones from OS (OSm27–34) were assembled into the OSm cluster. The source of lipolytic enzymes related to uncultured bacteria was shown in parenthesis. The topology of the tree was estimated by bootstrap analysis with 1,000 replicates. The bar indicates a 0.2 change per amino acid site.