Abstract
A total of 118 actinobacterial isolates were collected from the three types of termite nests (mound, carton, and subterranean nests) to evaluate their potential as a source of bioactive actinobacteria with antimicrobial activity. The highest number (67 isolates) and generic abundance (7 known genera) of actinobacterial isolates were obtained from carton nests. Streptomyces was the dominant genus in each type of termite nest. In the non-Streptomyces group, Nocardia was the dominant genus detected in mound and carton nests, while Pseudonocardia was the dominant genus in subterranean nests. A discovery trend of novel species (<99% similarity in the 16S rRNA gene sequence) was also observed in the termite nests examined. Each type of termite nest housed >20% of bioactive actinobacteria that could inhibit the growth of at least one test organism, while 12 isolates, belonging to the genera Streptomyces, Amycolatopsis, Pseudonocardia, Micromonospora and Nocardia, exhibited distinct antimicrobial activities. Streptomyces sp. CMU-NKS-3 was the most distinct bioactive isolate. It was closely related to S. padanus MITKK-103T, which was confirmed by 99% similarities in their 16S rRNA gene sequences. The highest level of extracellular antimicrobial substances was produced by the isolate CMU-NKS-3, which was grown in potato dextrose broth and exhibited a wide range (6.10×10−4–1.25 mg mL−1) of minimum inhibitory concentrations against diverse pathogens. We concluded that termite nests are an abundant source of bioactive strains of cultivable actinobacteria for future biotechnological needs.
Keywords: termite nest, Actinobacteria, functional diversity, 16S rRNA gene, bioactive compounds
Actinobacteria are Gram positive bacteria that possess a high guanine plus cytosine (G+C) content in their genomic DNA. They have been detected in diverse ecological niches. Their member species are known to be a main source of various bioactive compounds, accounting for 70% of all currently discovered antibiotics (7, 23). Among the antibiotics derived from actinobacteria, 75% are produced by Streptomyces spp. and 25% are produced by the non-Streptomyces group (known also as rare actinomycetes) (4). These antibiotics are produced industrially and mainly supplied for medical, pharmaceutical, and agricultural needs. On the other hand, the release of bioactive compounds to the environment through anthropogenic activity and horizontal gene transfer has led to the evolution of natural pathogens and increased drug resistance. Therefore, novel and potent bioactive compounds from novel microbial resources may overcome this pathogenic evolution. The discovery of actinobacteria from diverse and unexplored sources has also been linked to increased opportunities to obtain novel bioactive compounds (15, 34). We here aimed to explore the diversity of actinobacteria in termite nests and their potential as a source of bioactive compounds.
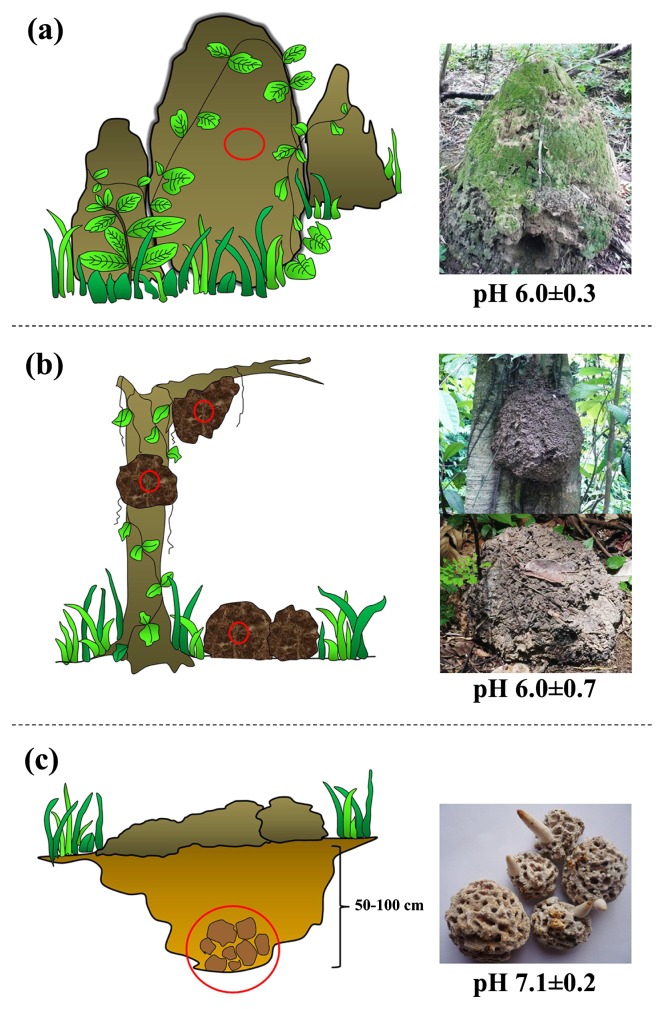
Termites are a group of social insects that are classified as the order Isoptera, which is closely related to cockroaches (5). They live in large colonies that comprise a king, queen, and workers, each of which have different behaviors and functions (19, 30, 38). Soil termites play an important role in circulating nutrients from decaying dead plant materials in the soil ecosystem (20, 29). Termite colonies are often found in a protective structure called a “termite nest”. These nests may be formed by diverse materials found close to their habitats. Mounds are a common type of termite nest, which are mainly constructed by the deposition of clay particles, organic carbon, and the saliva or secretions of the termites themselves. Other less common types, such as carton and subterranean nests, are also nest structures. Carton nests are mainly made from plant materials typically found on trees or in rich plant litter areas. The subterranean type is commonly found underground at an approximate depth of 50–100 cm. The various termite nests are shown in Fig. 1.
Fig. 1.
Diagram showing different types of termite nests. Three types of termite nests: mound (a), carton (b), and subterranean (c) nests, are indicated together with their average pH ± SD.
Recent studies reported no significant overlap in the gut microbial biota of soil-feeding termites and their nests or soil surrounding their nests (10, 11). Previous studies examined the microbial biota within the termite gut, and actinobacteria were identified as one of the dominant bacteria in this symbiotic lifestyle (3, 6, 20, 21, 26, 27). These symbiotic actinobacteria provide assisting functions for termites, such as nutrient cycling and exchange, and also protect termites from invading pathogens. Some of these termite-associated actinobacteria may also exhibit lignin-cellulolytic activity (3, 26, 27) and antagonistic activity against diverse pathogens (20, 21). Furthermore, some novel and prospective novel species of actinobacteria associated with termites have been reported previously (25, 32, 33). In the present study, we aimed to evaluate the generic and functional diversities of cultivable actinobacteria obtained from different types of termite nests. The biotechnological potential of the antimicrobial activity of actinobacteria was also evaluated against a set of representative pathogens found in medical and agricultural fields. The implication of different termite nests as an optimal source for potent antimicrobial actinobacteria was also discussed. The influences of some nutrition factors on the productivity of antimicrobial substances by a key actinobacterium and its further development for biotechnological applications were also assessed here.
Materials and Methods
Collection and preparation of termite nest samples
Three different termite nests, including mound, carton, and subterranean nests (Fig. 1), were collected from tropical forest and rubber tree farming areas in Phayao province, Thailand. Termite nest samples (200 g per sample from each type of termite nest) were placed in plastic bags and kept in an ice-box (for a maximum of 24 h) for transfer to the laboratory. All samples were ground into fine particles and air-dried at an ambient temperature for 7 d. Dried samples were suspended in sterilized distilled water and mixed well prior to the measurement of their pH by a pH meter (PB-10 Sartorius).
Isolation of actinobacteria from termite nests
Dried samples (10 g each) were pretreated either by moist heating in a water bath at 50°C for 1 h (35) or by suspending with 1.5% (w/v) phenol (15). These pretreatments were performed with the aim of reducing contamination by unwanted microorganisms (low G+C and/or Gram negative bacteria). Pretreated samples were serially diluted 10-fold, and the appropriate dilutions (10−3 to 10−6) were individually spread on starch casein (SC) agar (2) or humic acid-vitamin (HV) agar (14) supplemented with nystatin, cycloheximide, and nalidixic acid at final concentrations of 100, 100, and 50 μg mL−1, respectively. All plates were incubated at 30°C for 4 weeks. The individual colonies that appeared were re-grown and sub-cultured on International Streptomyces Project Medium 2 (ISP2) agar (2) at 30°C until they were pure isolates. Pure isolates were then kept either on Hickey-Tresner agar (2) as a working stock or in 20% (v/v) glycerol at −20°C for long term storage at the Department of Biology, Faculty of Science, Chiang Mai University.
Screening of antimicrobial actinobacteria isolated from termite nests
The antimicrobial activity of the actinobacterial isolates was evaluated by a dual culture technique against a list of test organisms indicated in Table 1. Antifungal activity against 5 phytopathogenic fungi was assessed on potato dextrose agar (PDA) (Kemma, Thailand). Briefly, an agar plug (5 mm Ø) of a test fungus grown previously on PDA at 30°C for 5 d was placed at the center of a new PDA plate, on which 2 lines (5 cm in length) of an actinobacterial isolate were individually streaked 3 cm from the fungal plug. The dual culture was incubated at 30°C for 7–14 d, and inhibition activity was observed intermittently. The plates inoculated with the fungi only served as controls. The antibacterial and anti-yeast activities of the actinobacterial isolates were assessed on nutrient agar (NA) (2) and Sabouraud agar (SBA) (2), respectively. Briefly, each actinobacterial isolate was streaked linearly at the center of each agar plate and allowed to grow at 30°C until mature colonies appeared (after approximately 3–5 d). The test organisms were streaked perpendicularly on their respective medium around a linear colony of the actinobacterial isolate. The dual culture was incubated at 37°C for the test bacteria used and at 30°C for the test yeasts used. The inhibition zone (a clear distance between the colonies of test organisms and actinobacterial isolate) that appeared was measured after 24 and 48 h of incubation. Control plates without the inoculation of actinobacteria were prepared to assess the normal growth of the test organisms on their respective agar. Distinctive antimicrobial isolates of the actinobacteria were selected based on their potential to inhibit a board range of the test organisms with relatively large inhibition zones. Of these isolates, the most distinctive antimicrobial isolate was chosen by its greater potential than that of the other isolates.
Table 1.
MIC values of a crude antimicrobial extract derived from Streptomyces sp. CMU-NKS-3.
| Test organism | MIC (mg mL−1) | ||
|---|---|---|---|
|
| |||
| Crude extract from Streptomyces sp. CMU-NKS-3 | Positive control | ||
|
| |||
| Streptomycin | Benomyl | ||
| Gram positive bacteriaa | |||
| Bacillus cereus1 | 1.22×10−3 | 0.02 | ND |
| Enterococcus faecalis ATCC 292172 | 0. 31 | 0.04 | ND |
| Micrococcus luteus3 | 1.22×10−3 | 2.44×10−3 | ND |
| Staphylococcus aureus ATCC 292134 | 2.44×10−3 | 4.88×10−3 | ND |
| Methicillin-resistant Staphylococcus aureus (MRSA)5 | — | 0.62 | ND |
| Gram negative bacteriab | |||
| Enterobacter aerogenes6 | 0.16 | 0.04 | ND |
| Escherichia coli ATCC 352187 | — | 0.08 | ND |
| Escherichia coli O157:H78 | — | 0.16 | ND |
| Klebsiella pneumoniae (ESBL+)9 | — | 0.04 | ND |
| Proteus mirabilis10 | 0.02 | 0.08 | ND |
| Proteus vulgaris11 | — | 0.02 | ND |
| Pseudomonas aeruginosa ATCC 2785912 | — | 0.08 | ND |
| Pseudomonas fluorescens13 | 0.04 | 2.50 | ND |
| Salmonella sp. Group D14 | — | 9.76×10−3 | ND |
| Salmonella typhae15 | — | 0.62 | ND |
| Serratia marcescens16 | — | 0.16 | ND |
| Yeastsc | |||
| Candida albicans17 | 6.10×10−4 | 0.31 | ND |
| Cryptococcus neoformans18 | 6.10×10−4 | 0.08 | ND |
| Filamentous fungid | |||
| Aspergillus flavus19 | 1.25 | ND | 0.31 |
| Colletotrichum musae20 | 0.62 | ND | 0.08 |
| Fusarium solani21 | 0.31 | ND | 0.08 |
| Rhizoctonia solani AG-222 | 0.16 | ND | 0.02 |
| Sclerotium solani23 | 0.16 | ND | 0.04 |
The initial stocks were kindly provided by the Central and Diagnostic Laboratory, Maharaj Nakorn Chiang Mai Hospital, Faculty of Medicine, Chiang Mai University, Thailand.
The filamentous fungi (phytopathogens) were taken from a culture collection at the Sustainable Development of Biological Resources Lab (SDBR), Department of Biology, Faculty of Science, Chiang Mai University.
Index numbers refer to the test organisms shown in Fig. 6. Not determined (ND) and no inhibition activity (—) were also observed, while methanol was used as a negative control for all tests. Initial concentrations of the crude extract and positive controls together with their dilution factors can be found in the Materials and Methods section. Tests were performed in triplicate, and no significant difference was observed in the results derived from triplicate experiments.
Diversity study of actinobacteria isolated from termite nests
Primary identification.
The generic diversities of all actinobacteria isolated from termite nests were determined and classified using some key phenotypes from the database available in the Bergey’s Manual of Systemic Bacteriology (40). The arrangement of spores (chain or single) was examined microscopically using a cover slip-implanting technique. The cover slip was implanted on actinobacteria-growing ISP2 agar and incubated at 30°C for 7–10 d, which allowed aerial mycelia to grow on the cover slip. The cover slip was removed from the culture plate, on which the existing mycelia were then fixed by 15.5% (v/v) acetic acid for 1 min. The fixed mycelia were observed under a light microscope (Olympus SZ40, Japan) after simple staining with crystal violet. The colony morphology of the actinobacteria grown on ISP2 agar was also observed under a stereomicroscope (Olympus CH30, Japan). An analysis of 2,6-diaminopimelic acid (DAP) was carried out to determine the cell wall chemotype of the actinobacteria, following the protocol described by Ningthoujam et al. (24). The isomeric type of DAP together with the colony morphology were used to divide non-Streptomyces and unidentified genera from the Streptomyces group. The unidentified genera were determined according to the limited formation of aerial mycelia and spores on the agar media used. Their growth on the liquid medium used in the protocol to analyze the cell wall chemotype was also restricted.
Secondary identification.
Only distinctive antimicrobial isolates of actinobacteria were selected to determine their phylogenetic position. The genomic DNA of these isolates was extracted from their biomass, which was obtained after their massive growth in ISP2 broth at 30°C. The extraction was performed following the manufacturer’s instructions for the FavorPrep™ Tissue Genomic DNA Extraction Mini Sample Kit (FAVORGEN®, Taiwan). The concentration and purity of DNA samples were evaluated by electrophoresis on a 1% (w/v) agarose gel and visualized by staining with 0.5 μg mL−1 ethidium bromide. An appropriate concentration of the DNA samples was adjusted for PCR amplification of the 16S rRNA gene. Taq DNA polymerase and a pair of universal primers (forward primer 27 (5′-AGAGTTTGATCCTGGCTCAG-3′) and reverse primer 1,525 (5′-AAGGAGGTGWTCCARCC-3′ (R=A/G, W=A/T)) were used (18). PCR was performed according to the steps described by Adegboye and Babalola (1). PCR products were purified using the NucleoSpin® Gel and PCR Clean-up Kit (MACHEREY-NAGEL, Germany) following the manufacturer’s instructions. Purified PCR products were sequenced using the facilities of First Base Laboratories, Malaysia. Almost-complete 16S rRNA gene sequences (approximately 1,500 nt) were determined and compared with the corresponding sequences available in the GenBank database (http://www.ncbi-nlm-nih.gov/) using the BLAST program. A multiple sequence alignment was carried out before the construction of a phylogenetic tree by MEGA program version 5 (36) with the neighbour-joining method (31). In pairwise comparisons, sequence similarities were computed using PHYDIT program version 1.0 (http://plaza.snu.ac.kr/jchun/phydit/).
Effects of different fermentation media on the production of antimicrobial compounds
The most distinctive antimicrobial isolate was chosen and evaluated for its potential to form antimicrobial substances. Six different fermentation broths including potato dextrose broth (PDB) (Kemmar, Thailand), YpSs (2), AMHU-5 (22), Emerson’s Broth (EM) (2), Bennett’s Broth (BN) (2), and F-4 (8) were used to grow the isolates. Seventy milliliters of each broth was used at 30°C and shaking at 125 rpm, for 7 d. The biomass was then removed by centrifugation at 6,000 rev min−1 for 15 min. The supernatant of each culture broth was extracted by ethyl acetate at a ratio of 1:1 (v/v). The mixture was mixed vigorously for 5 min and left overnight, and the extraction was repeated 3 times. The ethyl acetate residual was removed by evaporation at 40°C using a rotary vacuum evaporator (Rotavapors® R-215 BUCHI). The residual of antimicrobial substances was re-suspended in methanol at a concentration of 50 mg mL−1 and used as a crude extract for the antimicrobial test using the agar well diffusion assay against the same set of test organisms listed in Table 1.
Test bacteria were grown in nutrient broth (Difco™, USA) at 37°C and shaking at 125 rpm for 1 d, while the test yeasts were grown in Sabouraud broth (24) at 30°C and shaking at 125 rpm for 16 h, before use. The cell densities of both test organisms were then adjusted relative to the 0.5 McFarland Standard, which corresponded to 108 CFU mL−1. Spore suspensions of phytopathogenic fungi were prepared by adding 0.85% (w/v) NaCl solution and scraping the fungal mycelia previously grown on PDA for 3–5 d. The concentration of the fungal spores was adjusted by the NaCl solution to 1–3×106 spores mL−1 and quantified by a hemocytometer (BLAUBRAND®, Germany). All prepared suspensions of the test organisms were swabbed on their respective agar media including NA (for the test bacteria), SBA (for the test yeasts), and PDA (for the phytopathogenic fungi), on which wells were made. Fifty μL of the crude antimicrobial extract was added onto each agar well, while the same volumes of a negative control (methanol) and two positive controls (streptomycin [Wako Pure Chemical Industries, Japan] and benomyl [Banlee®, Thailand]) were added individually to the other wells. The positive controls were prepared by dissolving a powder of the antibiotics with methanol at the same concentration (50 mg mL−1) of the crude extract before use. Inhibition activity was observed by the appearance of inhibition zones (mm Ø), the sizes of which were recorded after an incubation at 37°C for 1 d (for the test bacteria), at 30°C for 1 d (for the test yeasts), and at 30°C for 3–5 d (for the phytopathogenic fungi). All tests were performed in triplicate.
Determination of minimal inhibitory concentrations (MICs)
The most distinctive antimicrobial isolate was grown in 500 mL of PDB at 30°C and shaking at 125 rpm for 7 d. Its cell-free culture fluid was prepared by filtration through Whatman® No. 1 filter paper, and was then extracted by ethyl acetate following a previous extraction protocol. The crude extract (5 mg mL−1) was serially diluted 2-fold, and appropriate dilutions (2.50 to 6.10×10−4 mg mL−1) were tested for their antimicrobial activity using the paper disc diffusion assay against the same set of test organisms listed in Table 1. Fifty μL of each dilution was loaded onto a sterilized filter paper disc, while the same volumes of negative control (methanol) and positive controls (streptomycin and benomyl) were also individually loaded onto the other paper discs. The positive controls were prepared by dissolving the powder of the antibiotics with methanol at the same concentration (5 mg mL−1) of the crude extract, while a set of 2-fold dilutions was performed as described above before use. MICs were determined as the lowest concentration showing inhibitory activity (appearance of an inhibition zone) against the test organisms. All tests were performed in triplicate.
Statistical analysis
Differences in means were evaluated by a one-way analysis of variance (one-way ANOVA) using Tukey’s post hoc at a significant level of P≤0.05 within SPSS version 17.0. The statistically analyzed data of F-distribution values were shown elsewhere in this study. The mean values with standard deviations (SDs) were computed on the basis of an at least triplicate data set.
Results
Termite nest actinobacteria and their generic abundance
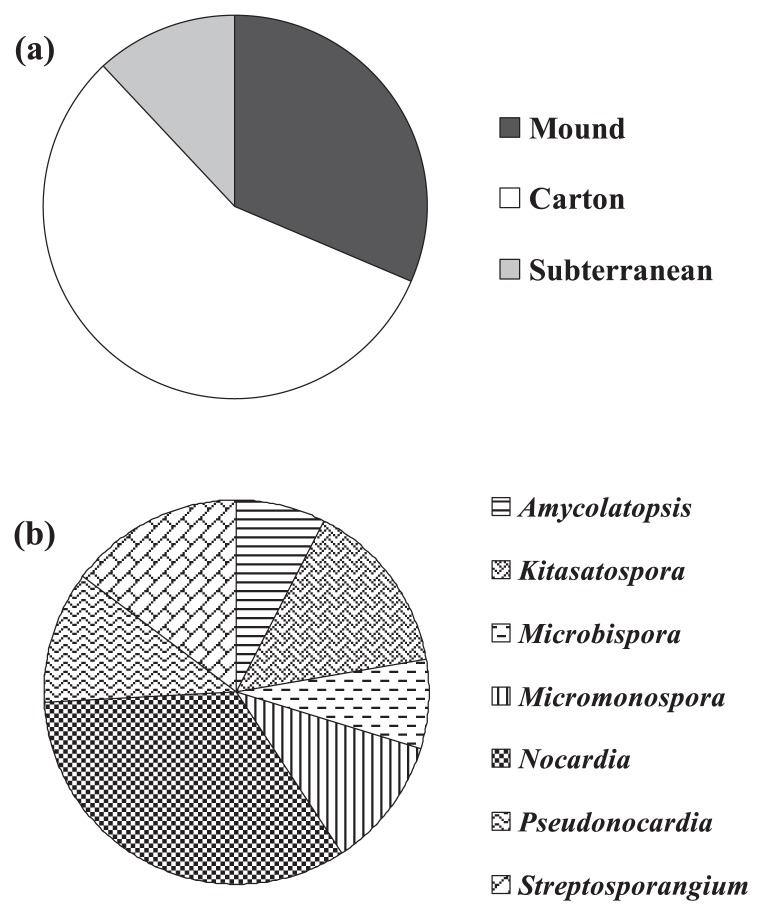
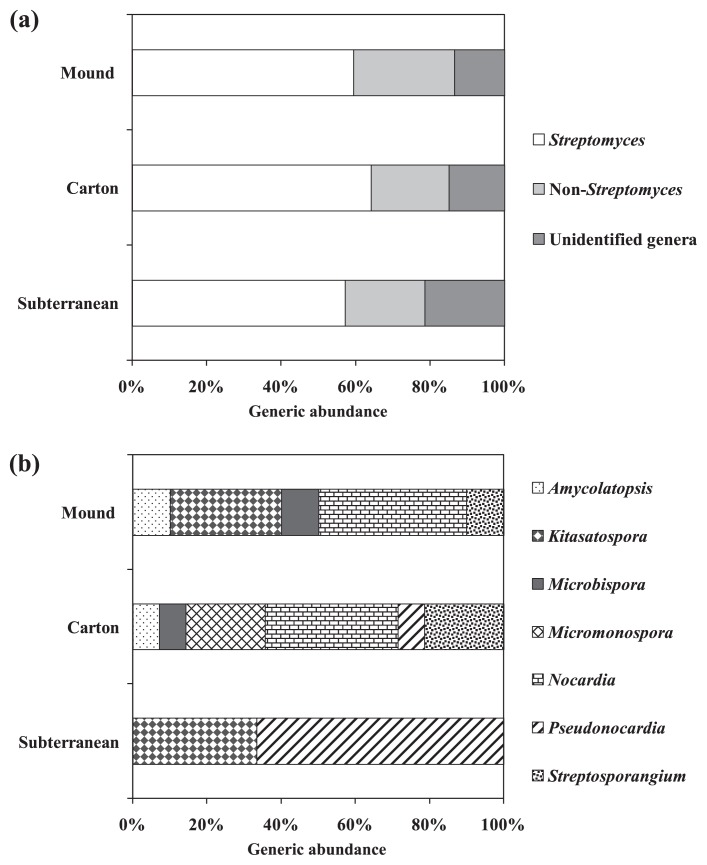
A total of 118 actinobacterial strains were isolated from 8 termite nest samples (Table 2), while HV agar with a pretreatment of 1.5% (w/v) phenol was the optimal isolation process, giving the highest number of isolates (44.9%). A range of termite nest pH (5.3–7.2) was measured, and the average pH in each termite nest was shown in Fig. 1. Forty-five actinobacterial isolates were derived from termite nest pH 5.3–5.8, 54 isolates from pH 6.0–6.6, and 19 isolates from pH 6.6–7.2. Among all isolates, 56.8% were derived from carton nests, 31.4% were derived from mound nests, and 11.9% were derived from subterranean nests (Table 2 and Fig. 2a). All isolates within the non-Streptomyces group (Table 2 and Fig. 2b) were primarily identified using key phenotypic data into 7 different genera including Amycolatopsis (7.4%), Kitasatospora (14.8%), Microbispora (7.4%), Micromonospora (11.1%), Nocardia (33.3%), Pseudonocardia (11.1%) and Streptosporangium (14.8%). The 16S rRNA gene sequencing data of some bioactive isolates belonging to the non-Streptomyces group confirmed that the primary identification was accurate. For example, isolates CMU-NKS-2, CMU-NKS-51, CMU-NKS-70, and CMU-NKS-77 were primarily identified as the genera Amycolatopsis, Micromonospora, Pseudonocardia, and Nocardia, respectively. Consequently, the phylogenetic positions of these isolates were determined to be closely related to Amycolatopsis mediterranei NRRL B-3240T, Micromonospora echinofusca HBUM 175187T, Pseudonocardia oroxyli D10T, and Nocardia harenae WS-26T, respectively (Table 3). In addition, slight differences were observed in the % abundance of the actinobacterial groups (Streptomyces, non-Streptomyces, and unidentified genera) found in each type of termite nest (Table 2 and Fig. 3a). However, Streptomyces was the dominant genus found in every type of termite nest. The generic abundance of the non-Streptomyces group varied across the different types of termite nests examined, and the most diverse genera (6 genera) were detected in carton nests and the fewest (2 genera) in subterranean nests (Table 2 and Fig. 3b). Among this non-Streptomyces group, Nocardia was the dominant genus found in mound and carton nests, while Pseudonocardia was the dominant genus found in subterranean nests.
Table 2.
Actinobacteria isolated from termite nests
| Termite nests | Streptomyces | Non-Streptomyces* | Unidentified genera | Total | Number of bioactive isolates that antagonized at least one test organism† | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||
| a | b | c | d | e | f | g | h | i | j | k | ||||
| Mound (3) | 22 | 1 | 3 | 1 | 0 | 4 | 0 | 1 | 5 | 37 | 3 | 6 | 2 | 3 |
| 5 | 8 | 4 | 5 | |||||||||||
| 3 | 4 | 3 | 6 | |||||||||||
|
| ||||||||||||||
| Carton (3) | 43 | 1 | 0 | 1 | 3 | 5 | 1 | 3 | 10 | 67 | 9 | 11 | 8 | 7 |
| 3 | 7 | 3 | 3 | |||||||||||
| 5 | 4 | 4 | 6 | |||||||||||
|
| ||||||||||||||
| Subterranean (2) | 8 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 3 | 14 | 2 | 4 | 1 | 4 |
| 2 | 3 | 2 | 1 | |||||||||||
|
| ||||||||||||||
| Total | 73 | 2 | 4 | 2 | 3 | 9 | 3 | 4 | 18 | 118 | 32 | 47 | 27 | 35 |
The numbers in parentheses refer to the number of termite nest samples.
The non-Streptomyces group comprised (a) Amycolatopsis, (b) Kitasatospora, (c) Microbispora, (d) Micromonospora, (e) Nocardia, (f) Pseudonocardia, and (g) Streptosporangium.
Test organisms comprised (h) Gram positive bacteria, (i) Gram negative bacteria, (j) yeasts, and (k) filamentous fungi.
Fig. 2.
Ratio of actinobacteria derived from each type of termite nest (a) and primary identification of actinobacteria that belong to the non-Streptomyces group (b). This identification was performed on the basis of some key phenotypic characterizations (see also the Materials and Methods section).
Table 3.
Phylogenetic analysis based on the 16S rRNA gene sequences of distinctive antimicrobial actinobacteria isolated from different types of termite nests
| Isolate number | Termite nests | Sequence length (nt) | Closest related bacteria in the GenBank database | Accession number | % Sequence similarity |
|---|---|---|---|---|---|
| CMU-NKS-2 | Carton | 1,239 | Amycolatopsis mediterranei NRRL B-3240T | KF746333 | 99 |
| CMU-NKS-3 | Carton | 1,375 | Streptomyces padanus MITKK-103T | KF746332 | 99 |
| CMU-NKS-5 | Subterranean | 1,435 | Streptomyces bungoensis NBRC 15711T | KF746334 | 98 |
| CMU-NKS-7 | Carton | 1,340 | Streptomyces hygroscopicus subsp. hygroscopicus NBRC 340T | KF746335 | 99 |
| CMU-NKS-12 | Mound | 1,359 | Streptomyces bingchengensis HBUM 174849T | KF746336 | 99 |
| CMU-NKS-51 | Carton | 1,345 | Micromonospora echinofusca HBUM 175187T | KF746341 | 99 |
| CMU-NKS-67 | Mound | 1,175 | Streptomyces roseocinereus NBRC 13829T | KF746337 | 99 |
| CMU-NKS-70 | Subterranean | 1,342 | Pseudonocardia oroxyli D10T | KF746342 | 99 |
| CMU-NKS-77 | Carton | 1,356 | Nocardia harenae WS-26T | KF746343 | 99 |
| CMU-NKS-79 | Mound | 1,328 | Streptomyces griseocarneus NBRC 13428T | KF746338 | 99 |
| CMU-NKS-110 | Mound | 1,071 | Streptomyces ginsengisoli Gsoil 025T | KF746339 | 99 |
| CMU-NKS-111 | Subterranean | 1,330 | Streptomyces bingchengensis HBUM174849T | KF746340 | 99 |
Fig. 3.
Generic diversity of actinobacteria isolated from each type of termite nest. All actinobacteria found in each type of termite nest were classified into 3 groups including Streptomyces, non-Streptomyces, and unidentified genera (a), while members of the non-Streptomyces group were primarily identified into their genera (b). This identification was performed on the basis of some key phenotypic characterizations (see also the Materials and Methods section).
Antimicrobial actinobacteria and their phylogenetic analysis
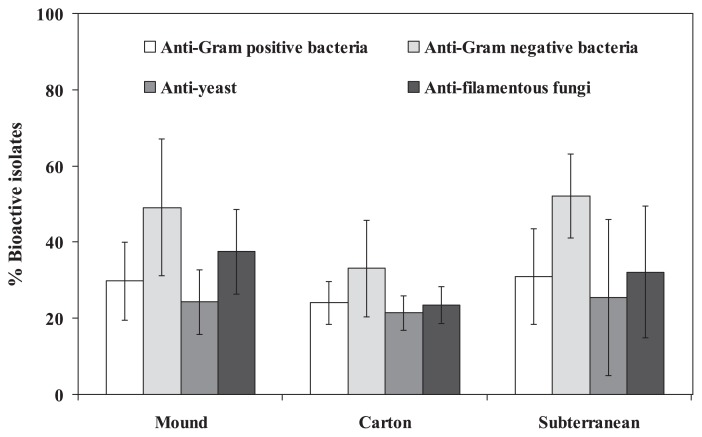
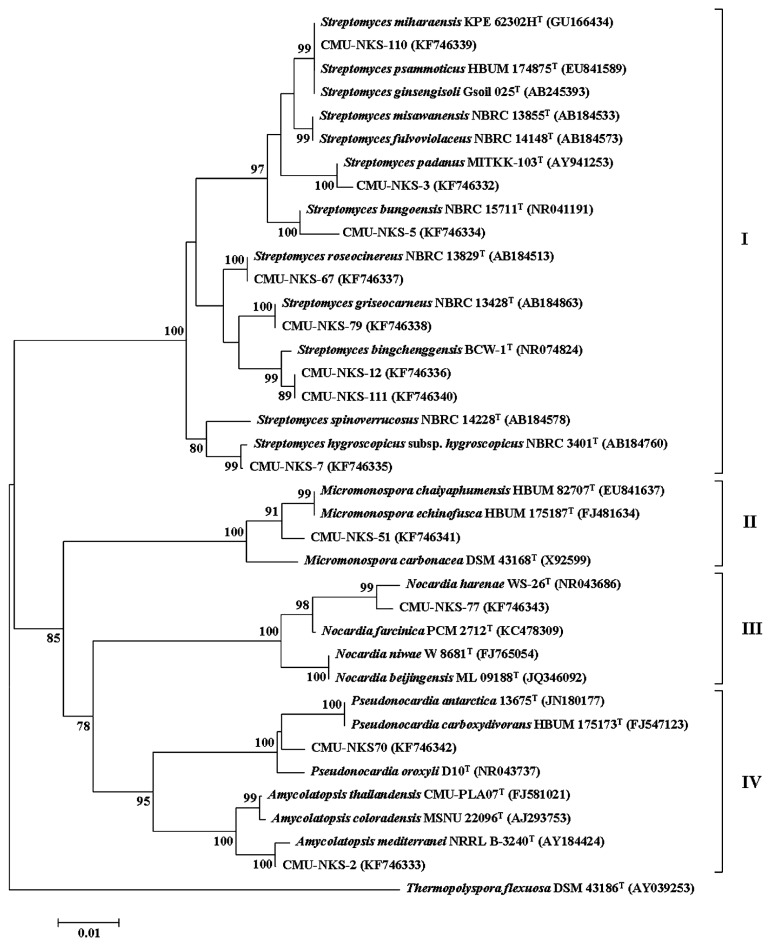
Among all the actinobacterial isolates examined, 47 isolates (39.8%) exhibited antibacterial activity, while 35 isolates (29.7%) exhibited antifungal activity. Most of these bioactive isolates were identified as Streptomyces, while a few isolates belonged to the non-Streptomyces group (Table 3). When we compared the abundance of the bioactive isolates derived from each type of termite nest, at least 20% of each could inhibit at least one test organism (Table 2 and Fig. 4), and most of the isolates could inhibit at least one Gram negative bacterium. However, no significant differences were observed in the % of bioactive isolates detected in the different types (mound, carton and subterranean) of termite nests or in the inhibition of different test organisms (Gram positive bacteriaF1, Gram negative bacteriaF2, yeastsF3, and filamentous fungiF4) (F1-4(2,5) = 0.429, 1.301, 0.095, 1.261, P=0.001). Among all bioactive actinobacteria, the distinctive antimicrobial activities of 12 isolates were greater than those of the other isolates, as listed in Table 3. Based on the phylogenetic analysis of these distinctive isolates, the % similarities of the 16S rRNA gene sequences matched to the closest sequences available in GenBank database were 99.0% except for the isolate CMU-NKS-5, which was closely related to Streptomyces bungoensis NBRC 15711T (98% sequence similarity, Table 3). Eight isolates belonged to the genus Streptomyces, while 4 isolates belonged to the non-Streptomyces genera, including Amycolatopsis (isolate CMU-NKS-2), Micromonospora (isolate CMU-NKS-51), Pseudonocardia (isolate CMU-NKS-70), and Nocardia (isolate CMU-NKS-77). The most distinctive antimicrobial isolate, Streptomyces sp. CMU-NKS-3 (which had the largest inhibition zone against most of the test organisms at the primary screening) was closely related to Streptomyces padanus MITKK-103T, as supported by 99% similarity in the 16S rRNA gene sequence. All distinctive isolates were classified into 4 families (Streptomycetaceae, Micromonosporaceae, Nocardiaceae and Pseudonocardiaceae) of the order Actinomycetales by phylogenetic analysis (Fig. 5), while the accession numbers (KF746332– KF746343) of their 16S rRNA gene sequences obtained after their deposition to the GenBank database can be found in Table 3.
Fig. 4.
Percentage of the bioactive isolates of actinobacteria isolated from each type of termite nest. The bioactive isolates were quantified based on their capacity to inhibit the growth of at least one test organism. The graph was plotted by means ± SDs computed based on the data derived from different population numbers (numbers of termite nest samples) taken from Table 2. Statistical comparisons can be found elsewhere in this study.
Fig. 5.
A neighbour-joining phylogenetic tree based on the 16S rRNA gene sequences, showing the relationship between the distinctive antimicrobial actinobacteria isolated from termite nests and recognized members of the class Actinobacteria. Thermopolyspora flexuosa DSM 43186T was used as an outgroup. Bootstrap values (>50%) based on 1000 replications are shown at the branch nodes. Bar = 0.01 substitutions per nucleotide position. The accession numbers of the gene sequences are indicated in parentheses. The phylogenetic tree was categorized into 4 families: (I) Streptomycetaceae, (II) Micromonosporaceae, (III) Nocardiaceae, and (IV) Pseudonocardiaceae.
Influence of different nutrients on the production of antimicrobial substances by Streptomyces sp. CMU-NKS-3
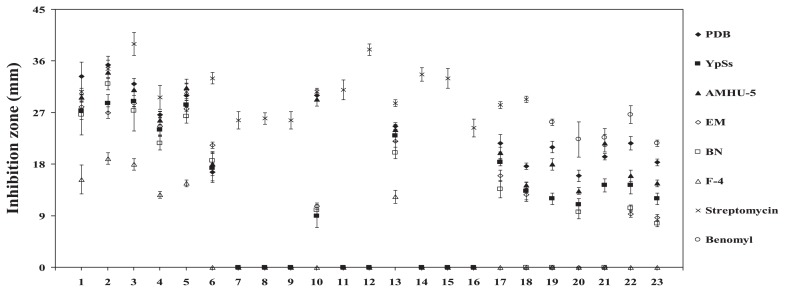
Isolate CMU-NKS-3 showed the most distinctive antimicrobial activity against most of the test organisms examined, and was assessed for its potential to form antimicrobial substances using 6 different fermentation broths (Fig. 6). Among all the broths tested, PDB and AMHU-5 were the optimal fermentation broths for the production of antimicrobial substances by the isolate, giving significant inhibitory activity that was equal to streptomycin or benomyl against Bacillus cereusF5, Enterococcus faecalis ATCC 29217F6, Staphylococcus aureus ATCC 29213F7, MRSAF8, Proteus mirabilisF9, and Fusarium solaniF10 (F5-10(6,14) = 23.828, 64.253, 51.056, 49.000, 415.597, 383.140, P=0.01). F-4 was the poorest fermentation broth for the production of antimicrobial substances by the isolate. Although AMHU-5 was one of the optimal fermentation broths, the crude extract derived from this broth showed significantly weaker antifungal activities against Cryptococcus neoformansF11, Aspergillus flavusF12, Rhizoctonia solani AG-2F1, and Sclerotium solaniF14 than those derived from PDB (F11-14(5,12) = 245.169, 570.600, 181.413, 311.886, P=0.01). Several Gram negative bacteria were able to resist the inhibitory activity of the crude extract derived from the isolate grown in any fermentation broth (Table 1 and Fig. 6). However, the isolate showed board ranges of antimicrobial activity against the test organisms (Gram positive and Gram negative bacteria, yeasts, and filamentous fungi). The MIC values (Table 1) of the crude extract derived from the isolate grown in PDB ranged from 6.10×10−4 to 1.25 mg mL−1, while the most susceptible test organisms were both yeasts (Candida albicans and Cryptococcus neoformans). However, most Gram negative bacteria and the Gram positive bacteria, methicillin-resistant Staphylococcus aureus (MRSA) were the strongest antagonists among all the test organisms; no inhibition activity was observed when any concentration of the crude extract was tested. The strongest antagonist among all phytopathogenic fungi was Aspergillus flavus, with a MIC value of 1.25 mg mL−1.
Fig. 6.
Effects of fermentation broths on the production of antimicrobial substances by Streptomyces sp. CMU-NKS-3. PDB, YpSs, AMHU-5, EM, BN, and F-4 were used as the fermentation broths (see also the Materials and Methods section). Streptomycin (for test bacteria and yeasts) and benomyl (for test filamentous fungi) at a final concentration of 50 mg mL−1 were used as the positive controls. The numbers 1–23 represent the test organisms listed in Table 1. The graph was plotted by means ± SDs computed based on the data derived from triplicate experiments. Statistical comparisons can be found elsewhere in this study.
Discussion
In the present study, termite nests were shown to be an abundant source of actinobacteria; 118 actinobacterial isolates could be obtained. These actinobacteria were classified into 8 known genera in 4 families of the order Actinomycetales, excluding some unidentified genera. The highest number (67 isolates) and generic abundance (7 known genera) among all actinobacterial isolates were detected in carton nests. A previous study revealed that actinobacteria were microbiota in mound termite nests constructed by Cubitermes niokoloensis in eastern Casamance (Senegal) (11). The optimal isolation process used for actinobacteria in termite nest samples was HV agar together with a pretreatment by phenol solution. HV agar supplemented with some antibiotics was previously reported to be an optimal selective medium for the isolation of actinobacteria from diverse soil habitats (9, 14, 16, 18). In addition, the pretreatment of samples prior to the isolation of actinobacteria is an important step that prevents contamination by low G+C and/or susceptible microorganisms that live in the same samples (13, 18). Termite nests constituted by different natural materials and different termite species lead to differences in the structural and physicochemical properties of the nests. This nest construction could be a selective ecological niche for actinobacteria. Our results showed that the pH of termite nests was slightly acid to neutral, while a higher abundance of actinobacteria was found in slightly acid termite nests than in neutral ones. We hypothesized that the location of termite nests may also influence the abundance of actinobacteria; a lower number of actinobacterial isolates was obtained from subterranean termite nests (found at a depth of 50–100 cm from the ground). This may be related to the oxygen limitation within this habitat because actinobacteria are aerobic microbes. In the present study, Streptomyces was the dominant genus found in every type of termite nest. However, a high generic abundance (8 genera) was also observed for the non-Streptomyces group, while the dominant genus within this group differed between the three termite nest types examined. Based on the results of our culture-dependent study, Nocardia (within the family Nocardiaceae) was the dominant genus found in carton and mound termite nests, which was similar to a metagenomic study of microbial biota living in mound termite nests reported by Fall et al. (11).
Previous studies demonstrated the biotechnological potentials of microbiota associated with termites. These microbes possess diverse bioactive functions to biosynthesize several applicable metabolites such as enzymes (3, 19, 26, 27, 29, 32) and antimicrobial substances (20, 21). Few studies have evaluated the antimicrobial function of cultivable actinobacteria isolated from termite-related sources especially in the termite gut (20, 21). We here demonstrated that termite nests housed abundant antimicrobial actinobacteria. Streptomyces has mainly been identified as the antagonist within termite-related habitats (20, 21), which was consistent with the results obtained in the present study. However, a high variety (4 genera within 3 families) of non-Streptomyces genera was also detected among the antimicrobial actinobacteria derived from the 3 termite nest types in this study. The ability to obtain novel actinobacteria that also exhibited antimicrobial activity was demonstrated. A previous study identified Saccharopolyspora pathumthaniensis sp. nov., a novel non-Streptomyces actinobacterium, in the termite gut. This novel species was closely related to Saccharopolyspora endophytica, as supported by 99.5% similarity in the 16S rRNA gene sequences, but a low DNA-DNA relatedness value of 53.3% (33). The 16S rRNA gene sequence generally needs to be examined to phylogenetically characterize microorganisms. However, the optimal range of this gene sequence similarity for determining differences in the genomic species varies depending on each phylogenetic species. The high sequence similarity (≥99%) of the gene, but low DNA-DNA relatedness (<70%) reported within the actinobacterial members is often found such as the example of the new species mentioned above. Therefore, recent studies (17, 28) assumed the novelty of actinobacterial isolates based on a lower % similarity in the gene sequences, approximately <99%, than their matched sequences available in the GenBank database. According to this predictive criterion, the actinobacterial isolate CMU-NKS-5, which showed 98% similarity in the gene sequence, could be assumed to be a novel species within the termite nest.
Different fermentation media were tested with the aim of evaluating the potential to form bioactive compounds of the most distinctive antimicrobial isolate, Streptomyces CMU-NKS- 3. In a recent evaluation, potato and dextrose in PDB were the optimal nitrogen and carbon sources for the production of antimicrobial agents by the isolate. Differences in carbon and nitrogen sources together with increasing temperatures are known to be an important factor for the production of bioactive compounds by the genus Streptomyces (12, 37). The second optimal medium (AMHU-5) for the isolate composed of 4% carbon source (glucose) and 3% nitrogen sources (soybean meal and yeast extract) was less suitable for the production of antifungal agents. This was in contrast to a combination of 5% glucose and 1% soybean meal, which was found to be an optimal medium for the production of antifungal agents by Streptomyces chattanoogensis (12). Therefore, not only the nutrient composition and growing conditions, but also genetic variations in their different genomic species influence the production of bioactive compounds by actinobacteria. Isolate CMU-NKS-3 was identified as Streptomyces padanus MITKK-103T, as supported by 99% sequence similarity in the 16S rRNA gene. Previous studies reported the potential to synthesize diverse antibiotics by S. padanus (39, 41). Recent studies by Wang et al. (39) and Ziong et al. (41) detected antifungal agents intracellularly within the mycelia of this Streptomyces species, while no antifungal activity was observed with the cell-free culture fluid. This was in contrast to our results in which antifungal activity was detected in the crude extract derived from the cell-free culture broth, which had relatively low MIC values against both single cell yeasts and phytopathogenic fungi (Table 1). This suggested that either the extracellular antifungal agents we gained may differ from the previously published compounds or isolate CMU-NKS-3 may be a novel species of the genus Streptomyces.
Conclusion
Termite nests represent an abundant source of cultivable actinobacteria that contribute diverse bioactive compounds for further applicable uses in the medical, pharmaceutical, and agricultural fields.
Acknowledgements
This work was supported by the Thailand Research Fund RTA 5580007, the Office of Higher Education Commission of Thailand under National Research University (A1) program, and the Graduate School, Chiang Mai University, Chiang Mai, Thailand. We also acknowledge Dr. Nareeluk Nakaew, the Department of Microbiology and Parasitology, Faculty of Medical Science, Naresuan University, Thailand, for her assistance in improving the phylogenetic analysis.
References
- 1.Adegboye MF, Babalola OO. Phylogenetic characterization of culturable antibiotic producing Streptomyces from rhizospheric soils. Mol. Biol. 2013;S1:001. [Google Scholar]
- 2.Atlas RM. Handbook of Microbiological Media. 2nd ed. CRC Press; Florida: 1946. [Google Scholar]
- 3.Basaglia M, Concheri G, Cardinali S, Pasti-Grigsby MB, Nuti MP. Enhanced degradation of ammonium-pretreated wheat straw by lignocellulolytic Streptomyces spp. Can J Microbiol. 1992;38:1022–1025. [Google Scholar]
- 4.Berdy J. Bioactive microbial metabolites. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 5.Berlanga M, Paster BJ, Guerrero R. The taxophysiological paradox: changes in the intestinal microbiota of the xylophagous cockroach Cryptocercus punctulatus depending on the physiological state of the host. Int Microbiol. 2009;12:227–236. [PubMed] [Google Scholar]
- 6.Bignell DE, Oskarsson H, Anderson JM. Association of actinomycetes-like bacteria with soil feeding termites (Termitidae, Termitinae) Appl Environ Microbiol. 1979;37:339–342. doi: 10.1128/aem.37.2.339-342.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler MS, Buss AD. Natural products—the future scaffolds for novel antibiotics. Biochem Pharmacol. 2006;71:919–929. doi: 10.1016/j.bcp.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 8.Cheeptham N, Higashiyama T, Phay N, Fukushi E, Matsuura H, Mikawa T, Yokota A, Ichihara A, Tomita F. Studies on an antifungal antibiotic from Ellisiodothis inquinans L1588-A8. Thai J Biotechnol. 1999;1:37–45. [Google Scholar]
- 9.Duangmal K, Ward AC, Goodfellow M. Selective isolation of members of the Streptomyces violaceoruber clade from soil. FEMS Microbiol Lett. 2005;245:321–327. doi: 10.1016/j.femsle.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 10.Fall S, Nazaret S, Chotte JL, Brauman A. Bacterial density and community structure associated with aggregate size fractions of soil-feeding termite mounds. Microb Ecol. 2004;28:191–199. doi: 10.1007/s00248-003-1047-2. [DOI] [PubMed] [Google Scholar]
- 11.Fall S, Hamelin J, Ndiaye F, Assigbetse K, Aragno M, Chotte JL, Brauman A. Differences between bacterial communities in the gut of a soil-feeding termite (Cubitermes niokoloensis) and its mounds. Appl Environ Microbiol. 2007;73:5199–5208. doi: 10.1128/AEM.02616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupte MD, Kulkarni PR. A study of antifungal antibiotic production by Streptomyces chattanoogensis MTCC 3423 using full factorial design. Lett Appl Microbiol. 2002;35:22–26. doi: 10.1046/j.1472-765x.2002.01119.x. [DOI] [PubMed] [Google Scholar]
- 13.Hayakawa H. Studies on the isolation and distribution of rare actinomycetes in soil. Actinomycetologica. 2008;22:12–19. [Google Scholar]
- 14.Hayakawa M, Nonomura H. Humic acid-vitamin agar, a new medium for selective isolation of soil actinomycetes. J Ferment Technol. 1987;65:501–509. [Google Scholar]
- 15.Hayakawa M, Yoshida Y, Iimura Y. Selection of bioactive soil actinomycetes belonging to the Streptomyces violaceusniger phenotype cluster. J Gen Appl Microbiol. 2004;96:973–981. doi: 10.1111/j.1365-2672.2004.02230.x. [DOI] [PubMed] [Google Scholar]
- 16.Hop DV, Sakiyama Y, Binh CT, Otoguro M, Hang DT, Miyadoh S, Luong DT, Ando K. Taxonomic and ecological studies of actinomycetes from Vietnam: isolation and genus-level diversity. J Antibiot. 2011;64:599–606. doi: 10.1038/ja.2011.40. [DOI] [PubMed] [Google Scholar]
- 17.Kaewkla O, Franco CMM. Rational approaches to improving the isolation of endophytic actinobacteria from Australian native trees. Microb Ecol. 2013;65:384–393. doi: 10.1007/s00248-012-0113-z. [DOI] [PubMed] [Google Scholar]
- 18.Khamna S, Yokota A, Lumyong S. Actinomycetes isolated from medicinal plant rhizosphere soils: diversity and screening of antifungal compounds, indole-3-acetic acid and siderophore production. World J Microbiol Biotechnol. 2009;25:649–655. [Google Scholar]
- 19.Khucharoenphaisan K, Puangpetch U, Puttaraksa K, Sinma K. Grouping of Actinomycetes isolated from termites using biochemical character. J Biol Sci. 2011;11:314–319. [Google Scholar]
- 20.Khucharoenphaisan K, Sripairoj N, Sinma K. Isolation and identification of actinomycetes from termite’s gut against human pathogen. Asian J Anim Vet Adv. 2012;7:68–73. [Google Scholar]
- 21.Matsui T, Tanaka J, Namihira T, Shinzato N. Antibiotics production by an actinomycete isolated from the termite gut. J Basic Microbiol. 2012;52:1–5. doi: 10.1002/jobm.201100500. [DOI] [PubMed] [Google Scholar]
- 22.Nakaew N, Pathom-aree W, Lumyong S. Generic diversity of rare actinomycetes from Thai cave soils and their possible use as new bioactive compounds. Actinomycetologica. 2009;23:21–26. [Google Scholar]
- 23.Newman DJ, Cragg MG. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 24.Ningthoujam DS, Sanasam S, Nimaichand S. Screening of actinomycete isolates from niche habitats in Manipur for antibiotic activity. Am J Biochem Biotechnol. 2009;5:221–225. [Google Scholar]
- 25.Ohkuma M, Kudo T. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:461–468. doi: 10.1128/aem.62.2.461-468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasti MB, Belli ML. Cellulolytic activity of actinomycetes isolated from termites (Termitidae) gut. FEMS Microbiol Lett. 1985;26:107–112. [Google Scholar]
- 27.Pasti MB, Pometto AL, 3rd, Nuti MP, Crowford DL. Lignin-solubilizing ability of actinomycetes isolated from termite (Termitidae) gut. Appl Environ Microbiol. 1990;56:2213–2218. doi: 10.1128/aem.56.7.2213-2218.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin S, Li J, Chen H-H, Zhao G-Z, Zhu W-Y, Jiang C-L, Xu L-H, Li W-J. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl Environ Microbiol. 2009;75:6176–6186. doi: 10.1128/AEM.01034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramin M, Alimon AR, Sijam K, Abdullah N. Filter paper degradation by bacteria isolated from local termite gut. Res J Microbiol. 2008;3:565–568. [Google Scholar]
- 30.Robert OE, Frank UO, Agbonsalo OU. Influence of activities of termites on some physical and chemical properties of soils under different land use patterns: a review. Int J Soil Sci. 2007;2:1–14. [Google Scholar]
- 31.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 32.Schafer A, Konrad R, Kuhnigk T, Kampfer P, Hertel H, Konig H. Hemicellulose-degrading bacteria and yeasts from the termite gut. J Appl Microbiol. 1996;80:471–478. doi: 10.1111/j.1365-2672.1996.tb03245.x. [DOI] [PubMed] [Google Scholar]
- 33.Sinma K, Ishida Y, Tamura T, Kitpreechavanich V, Tokuyama S. Saccharopolyspora pathumthaniensis sp. nov., a novel actinomycetes isolated from termite guts (Speculitermes sp.) J Gen Appl Microbiol. 2011;57:93–100. doi: 10.2323/jgam.57.93. [DOI] [PubMed] [Google Scholar]
- 34.Subramani R, Aalbersberg W. Marine actinomycetes: an ongoing source of novel bioactive metabolites. Microbiol Res. 2012;167:571–580. doi: 10.1016/j.micres.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Takizawa M, Colwell RR, Hell RT. Isolation and diversity of actinomycetes in the Chesapeake Bay. Appl Environ Microbiol. 1993;59:997–1002. doi: 10.1128/aem.59.4.997-1002.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theobald U, Schimana J, Fiedler HP. Microbial growth and production kinetic of Streptomyces antibioticus Tü 6040. Antonie van Leeuwenhoek. 2000;78:307–313. doi: 10.1023/a:1010282818272. [DOI] [PubMed] [Google Scholar]
- 38.Verma M, Sharma S, Prasad R. Biological alternatives for termite control: a review. Int Biodet Biodeg. 2009;63:959–972. [Google Scholar]
- 39.Wang YF, Wei SJ, Zhang ZP, Zhan TH, Tu GQ. Antifungalmycin, an antifungal macrolide from Streptomyces padanus 702. Nat Prod Bioprospect. 2012;2:41–45. [Google Scholar]
- 40.Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Suzuki K, Ludwig W, Whitmann WB. Bergey’s Manual of Systematic Bacteriology. 2nd ed. Vol. 5. Springer; New York: 2012. [Google Scholar]
- 41.Xiong ZQ, Zhang ZP, Li JH, Wei SJ, Tu GQ. Characterization of Streptomyces padanus JAU4234, a producer of actinomycin X2, fungichromin, and a new polyene macrolide antibiotic. Appl Environ Microbiol. 2012;78:589–592. doi: 10.1128/AEM.06561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]