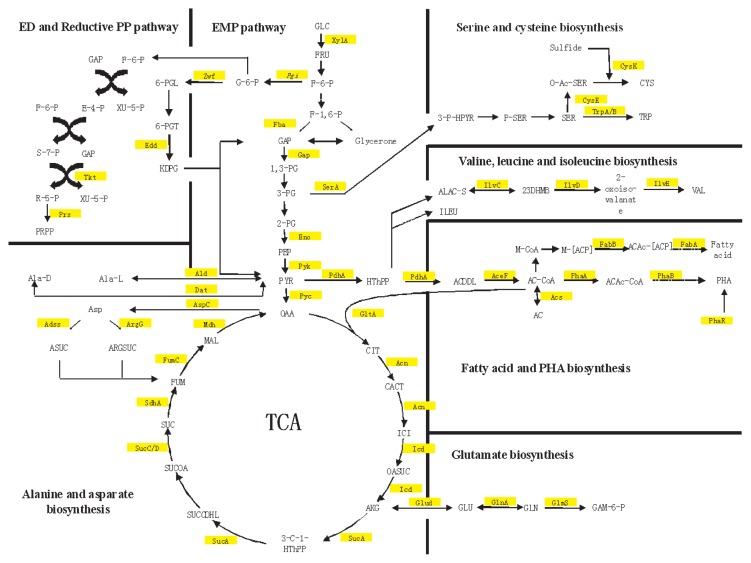
Fig. 2.
Central metabolic pathways of R. litoralis OCh149 in the stationary phase. Enzymes identified using LC-MS/MS are highlighted. Abbreviations (metabolites): GLC, glucose; FRU, fructose; F-6-P, fructose-6P; F-1,6-P, fructose-1,6P; GAP, glyceraldehyde-3P; 1,3-PG, glycerate-1,3P; 3-PG, glycerate-3P; 2-PG, glycerate-2P; PEP, phosphoenol-pyruvate; PYR, pyruvate; OAA, oxaloacetate; CIT, citrate; CACT, cis-aconitate; ICI, isocitrate; OASUC, oxalosuccinate; AKG, 2-oxo-glutarate; 3-C-1-HThPP, 3-carboxy-1-hydroxypropyl-ThPP; SUCCDHL, S-succinyldihydrolipoamide-E; SUCOA, succinyl-CoA; SUC, succinate; FUM, fumarate; MAL, malate; G-6-P, glucose-6P; 6-PGL, 6-phosphogluconolactone; 6-PGT, 6-phosphogluconate; KDPG, 2-dehydro-3-deoxygluconate-6P; E-4-P, erythrose-4P; XU-5-P, xylulose-5P; S-7-P, sedoheptulose-7P; R-5-P, ribose-5P; GLU, glutamate; GLN, glutamine; GAM-6-P, glucosamine-6P; Ala-L, L-alanine; Ala-D, D-alanine; Asp, asparatate; ARGSUC, arginino-succinate; ASUC, adenylo-succinate; 3-P-HPYR, 3P-hydroxy-pyruvate; P-SER, phosphoserine; SER, serine; O-Ac-SER, O-acetyl-serine; CYS, cysteine; TRP, L-tryptophan; ALAC-S, (s)-2-acetolactate; 23DHMB, (R)-2,3-dihydroxy-3-methylbutanoate; ILEU, isoleusine; VAL, valine; HTHPP, 2-hydroxyethyl-ThPP; ACDDL, S-acetyl-dihydrolipoamide-E; AC-CoA, acetyl-CoA; ACAc-CoA, acetoacetyl-CoA; M-CoA, malonyl-CoA; M-[ACP], malonyl-[acp]; ACAc-[ACP], acetoacetyl-[acp]; AC, acetate. Abbreviations (enzymes): XylA, xylose isomerase; Pgi, glucose-6-phosphate isomerase; Zwf, glucose-6-phosphate-1-dehydrogenase; Fba, fructose-bisphosphate aldolase; Gap, glyceraldehyde-3-phosphate dehydrogenase; Eno, enolase; Pyk, pyruvate kinase; Pyc, pyruvate carboxylase; Edd, phosphogluconate dehydratase; Tkt, transketolase; Prs, ribose-phosphate pyrophosphokinase putative; GltA, citrate synthase; Acn, aconitate hydratase; Icd, isocitrate dehydrogenase (NADP-dependent); SucA, alpha-ketoglutarate dehydrogenase; SucC, succinyl-CoA synthetase, beta subunit; SucD, succinyl-CoA synthetase, alpha subunit; SdhA, succinatedehydrogenase flavoprotein subunit; FumC, fumarate hydratase; Mdh, malate dehydrogenase; Ald, alanine dehydrogenase; Dat, D-alanine aminotransferase; AspC, aspartate aminotransferase; Adss, adenylosuccinate synthetase; ArgG, argininosuccinate synthase; CysE, serine acetyltransferase; CysK, O-acetylserine sulfhydrylase; TrpA, tryptophan synthase, alpha subunit; TrpB, tryptophan synthase, beta subunit; IlvC, ketol-acidreductoisomerase; IlvD, dihydroxy-acid dehydratase; IlvE, branched-chain amino acid aminotransferase; Glud, glutamate dehydrogenase; GlnA, glutamine synthetase, type I; GlmS, D-fructose-6-phosphate amidotransferase; PdhA, pyruvate dehydrogenase complex, E1 component, alpha subunit; AceF, dihydrolipoamide acetyltransferase; PhaA, acetyl-CoA acetyltransferase; PhaB, acetoacetyl-CoA reductase; PhaR, polyhydroxyalkanoate synthesis repressor; FabA, 3-hydroxydecanoyl-ACP dehydratase; FabB, 3-oxoacyl-(acyl-carrier-protein) synthase; Acs, acetyl-coenzyme A synthetase. Detailed information about the proteins is provided in supplemental material Table S1.