Table1.
Simulation study results
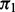 |
Enr. | Strat. | Rejected | FDP |
|---|---|---|---|---|
| 0.00 | None | None | 1 [0,5] | 1.00 [0.00,1.00] |
| 0.00 | None | Low | 4 [0,15] | 1.00 [0.00,1.00] |
| 0.001 | None | None | 79 [45,137] | 0.25 [0.11,0.42] |
| 0.001 | None | Low | 19 [4,70] | 0.55 [0.19,0.79] |
| 0.001 | Low | None | 92 [62,149] | 0.30 [0.00,0.46] |
| 0.001 | Low | Low | 17 [4,77] | 0.44 [0.00,0.70] |
| 0.001 | High | None | 90 [63,132] | 0.28 [0.13,0.41] |
| 0.001 | High | Low | 17 [5,47] | 0.46 [0.21,0.67] |
| 0.01 | None | None | 7 [1,19] | 0.00 [0.00,0.17] |
| 0.01 | None | Low | 6 [1,18] | 0.25 [0.00,0.85] |
| 0.01 | Low | None | 43 [17,101] | 0.10 [0.00,0.20] |
| 0.01 | Low | Low | 9 [1,38] | 0.23 [0.00,0.67] |
| 0.01 | High | None | 60 [16,124] | 0.11 [0.00,0.23] |
| 0.01 | High | Low | 8 [1,28] | 0.14 [0.00,1.00] |
| 0.05 | None | None | 4 [0,17] | 0.00 [0.00,0.17] |
| 0.05 | None | Low | 4 [0,15] | 0.00 [0.00,1.00] |
| 0.05 | Low | None | 39 [8,106] | 0.00 [0.00,0.07] |
| 0.05 | Low | Low | 8 [2,25] | 0.00 [0.59,0.23] |
| 0.05 | High | None | 47 [18,101] | 0.00 [0.00,0.07] |
| 0.05 | High | Low | 8 [1,27] | 0.00 [0.00,0.23] |
Note: Median number of SNPs rejected (Rejected) and FDP for the proposed cmfdr methodology. Settings include level of polygenicity (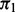 ), level of covariate enrichment (Enr.) and level of population stratification (Strat.). Numbers in brackets give middle 95% of distributions across 100 simulations for each setting. A SNP was rejected if its cmfdr was
), level of covariate enrichment (Enr.) and level of population stratification (Strat.). Numbers in brackets give middle 95% of distributions across 100 simulations for each setting. A SNP was rejected if its cmfdr was 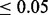 . Details of simulation settings and more extended comparisons are given in the Supplementary Materials.
. Details of simulation settings and more extended comparisons are given in the Supplementary Materials.
