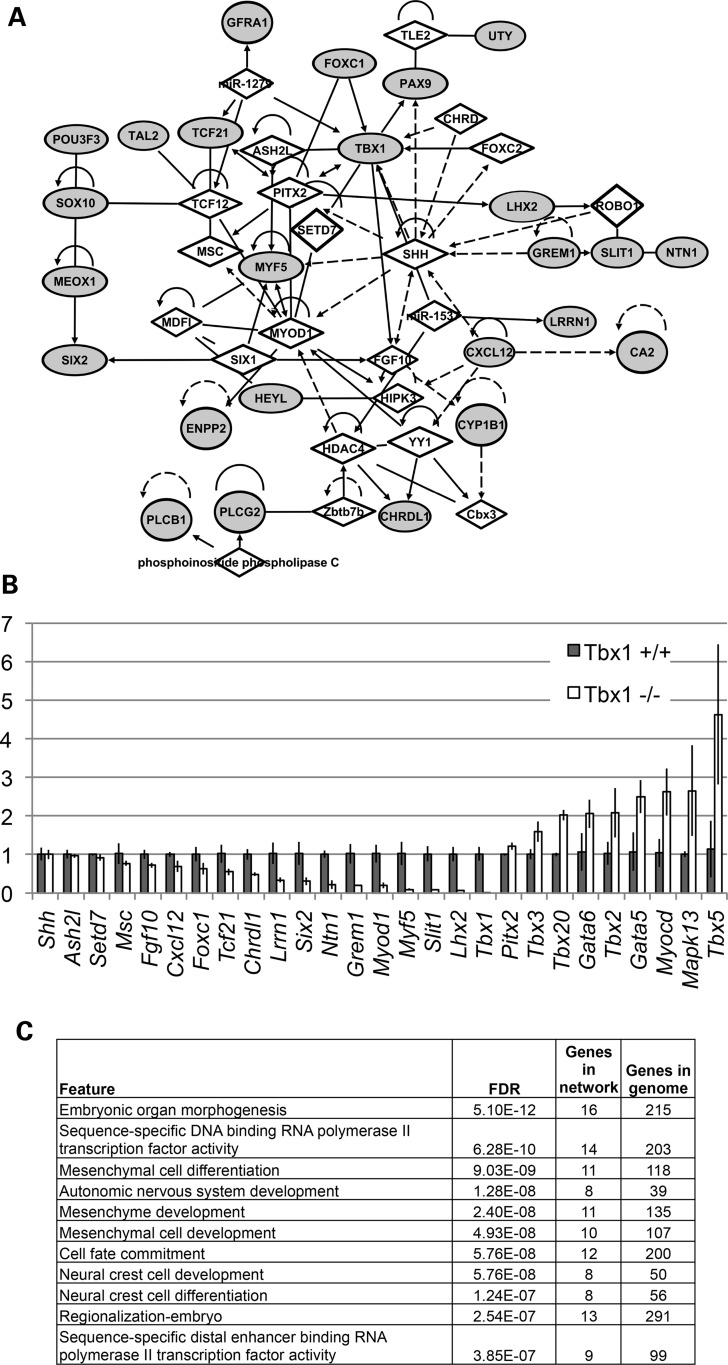
Figure 2.
Gene network of PA1 genes reduced in expression at E9.5 in Tbx1−/− embryos. (A) The network of genes that were reduced in expression in Tbx1−/− embryos when compared with Tbx1+/+ embryos (>1.5-fold change, P < 0.05) was created using IPA (http://www.ingenuity.com/products/ipa). The top network is depicted here. The genes reduced in expression are highlighted by gray fill, and additional genes connected to them based on IPA network analysis are indicated as diamonds. The lines indicate relationships between genes that could directly or indirectly interact. Genes reduced in expression but not linked to this network can be found in the Supplementary Material, Table S1. (B) Quantitative RT-PCR of PA1 tissue from Tbx1+/+ and Tbx1−/− embryos at E9.5. The X-axis indicates genes analyzed and Y-axis represents fold change of expression in PA1 tissue from Tbx1−/− relative to Tbx1+/+controls at E9.5. Error bars, s.e.m. (n = 3). (C) Genes reduced in expression in Tbx1−/− embryos at E9.5 were analyzed at the Genemania website (http://www.genemania.org/) to identify top enriched functions. We provide the FDR (false discovery rate) score estimated in Genemania by the Benjamini–Hochberg correction.