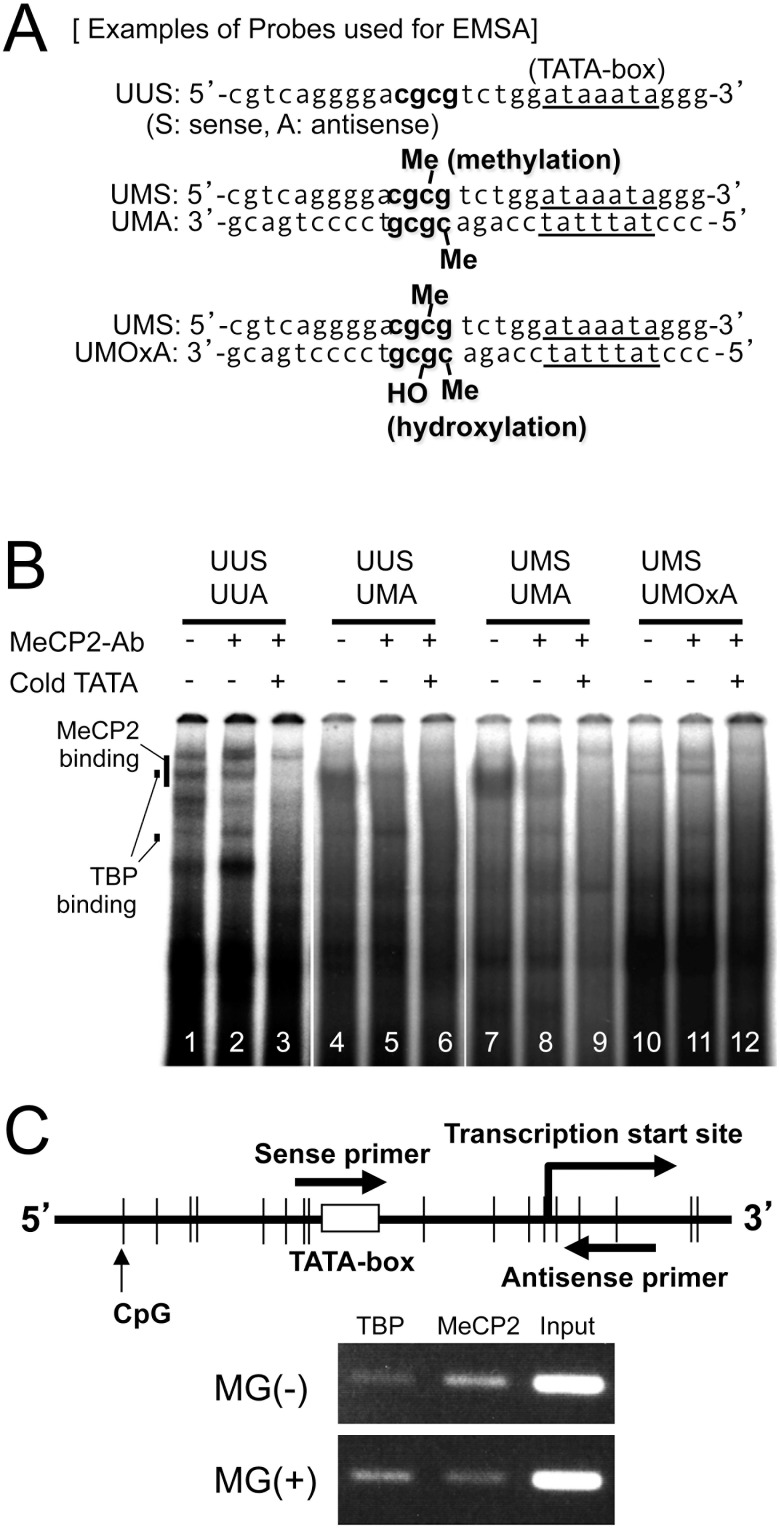
Figure 4. The effect of methylation and oxidation at CpG locus 5 bases upstream of TATA-box on MeCP2 binding.
In vitro binding of nuclear protein from HeLa cells to TATA-box and CpGs 5 bases upstream of TATA-box was tested by EMSA. (A) Double-stranded oligonucleotides, unmethylated (UUS/UUA), hemimethylated (UUS/UMA), single-methylated (UMS/UMA), and hemihydroxilated as well as single-methylated (UMS/UOxMA) ones, spanning part of the mouse sFRP-4 gene basic promoter region including TATA-box (−57/−29), were subjected to the binding reaction. (B) The unmethylated oligonucleotides show protein-DNA bindings (lanes 1 and 2, TBP binding) which are washed out by the cold consensus TATA-box sequence (lane 3). On the other hand, both hemi- and bi-methylated oligonucleotides show dense and clear protein-DNA binding (lanes 4 and 7, MeCP2 binding) which is block-shifted with an anti-MeCP2 antibody (lanes 5 and 8). At the same time, the protein-DNA bindings appearing at low and high positions (lanes 5 and 8, TBP binding) are washed out by a cold TATA-box competitor (lanes 6 and 9). By inducing a single 8-OHdG, protein-DNA bindings seen in lanes 4 and 7 have disappeared (lane 10), and alternative protein-DNA binding appearing at low and high positions (lanes 10 and 11, TBP binding) are washed out with a cold TATA-box competitor (lane 12). (C) A set of primers was used to amplify 80 bases of DNA containing a CpG dinucleotide, TATA-box and a transcription start site for ChIP assay of TBP and MeCP2 on the mouse sFRP-4 gene promoter. ST2 cells with or without MG treatment were subjected to immunoprecipitation. Input and immunoprecipitated DNA with either anti-TBP or anti-MeCP2 antibody was assessed by PCR using the set of primers as described above. Without MG treatment, a reactive PCR product reflecting mainly endogenous MeCP2 binding is observed. Conversely, a PCR product reflecting TBP binding was found mainly in MG-treated ST2 cells.