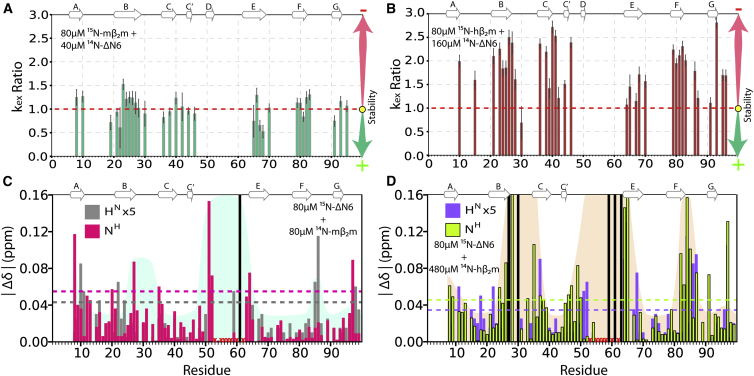
Figure 6.
Dynamic versus Rigid Body Interactions in Different Protein Complexes
(A) The ratio of the H/D exchange rates of 15N-labeled mβ2m bound (∼22%) to unlabeled ΔΝ6 (80 μΜ mβ2m + 40 μΜ ΔΝ6) versus free (80 μΜ) 15N-labeled mβ2m (kex ratio, bound:free) plotted against residue number. An increase in kex ratio indicates a loss of H/D exchange protection upon binding. Error bars represent the propagated error of the fits to the raw data shown in Figure S6.
(B) As in (A) but for free (80 μΜ) 15N-labeled hβ2m versus ∼22% 15N-labeled hβ2m bound to ΔΝ6 (80 μΜ hβ2m + 160 μΜ ΔΝ6). Note that exchange of hβ2m occurs by a mixed EX1/EX2 mechanism (Hodkinson et al., 2009), ruling out analysis of these data in terms of the free energy of binding.
(C) Differences in 1H (gray) and 15N (red) chemical shifts when 15N-labeled ΔΝ6 and 14N-labeled mβ2m are mixed in a 1:1 ratio (80 μΜ each; ∼45% ΔΝ6-bound). Dotted lines represent two standard deviations of the mean over the entire data set for each nucleus.
(D) As in (C) but for 15N-labeled ΔΝ6 mixed with 14N-labeled hβ2m (80 μΜ hβ2m + 480 μΜ ΔΝ6; ∼45% ΔΝ6-bound). Black bars denote residues that are broadened beyond detection but show significant chemical shift changes when less mβ2m/hβ2m is added. Red crosses denote ΔΝ6 residues that are broadened at pH 6.2. Residues that experience significant chemical shift changes on binding are highlighted in blue and pink backgrounds in (C) and (D), respectively.