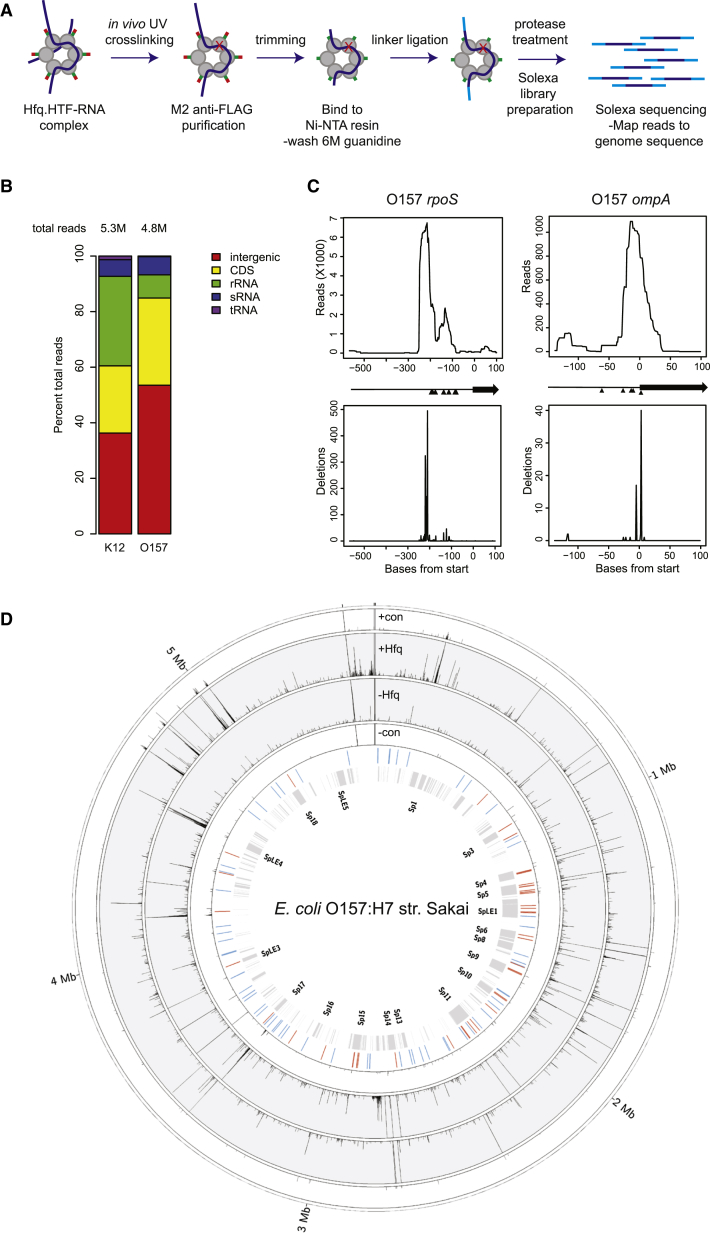
Figure 1.
UV Crosslinking of Hfq-RNA Correlates with In Vitro Footprinting of Hfq to Abundant mRNAs
(A) Workflow for CRAC analysis of Hfq. A detailed protocol is presented in Supplemental Experimental Procedures and Figure S1.
(B) Distribution of Hfq-bound reads between transcript classes in E. coli K12 str. MG1655 and E. coli O157 str. Sakai. Total reads are indicated above bars.
(C) Sequencing reads recovered from Hfq CRAC that map to rpoS or ompA mRNAs (top) and deletions recovered within sequencing reads (below). Black arrows between plots indicate the position of coding sequence (arrow) and 5′ UTR (line). Black triangles indicate position of nucleotides protected by Hfq in footprinting experiments in vitro (Moll et al., 2003; Soper and Woodson, 2008).
(D) Transcriptome-wide profiling of Hfq binding sites. Numbers of Hfq-associated reads mapped to the positive strand (+Hfq) and negative strand (−Hfq) are plotted in the gray line plots (y axis maximum 20,000 reads). Control experiments with untagged protein are plotted in the white outer and inner line plots (con±; y axis maximum 10,000 reads). From the inner-most track: text indicates designations for pathogenicity islands, with the position of all pathogenicity islands indicated by the gray boxes in the next track. The positions of sRNAs identified in this study are indicated in red, with previously described sRNAs in blue.