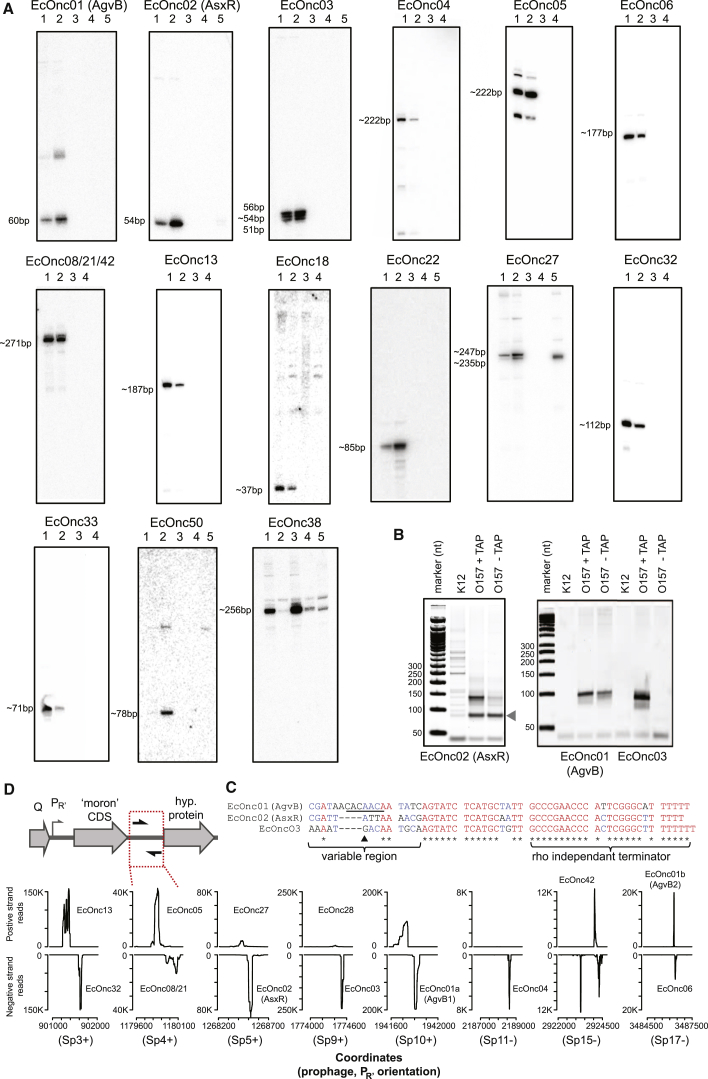
Figure 4.
Identification of Prophage-Encoded sRNAs in E. coli O157
(A) Northern blot analysis of predicted sRNAs (also see Table S2) in E. coli O157:H7 str. Sakai (O157) and nonpathogenic E. coli K12 (K12) cultured under virulence inducing conditions (MEM-HEPES) and in LB broth. Lane 1: O157 grown in MEM-HEPES; lane 2: O157 grown in LB; lane 3: K12 grown in MEM-HEPES; lane 4: K12 grown in LB; lane 5 (where applicable): O157Δhfq grown in LB. Approximate size of RNAs indicated left of blot.
(B) 5′ RLM-RACE with and without tobacco acid pyrophophatase (TAP) treatment of EcOnc01–EcOnc03. Grey arrow indicates a primer dimer.
(C) Prophage encode convergent sRNAs within the “moron” insertion site at PR’. (Top) Graphical representation of gene organization at the moron CDS insertion site showing the phage regulator, antiterminator Q CDS, and promoter PR’. Moron CDSs are inserted downstream of PR’, and convergent sRNAs are encoded between the moron CDS and a conserved hypothetical phage ORF. (Bottom) Hfq-bound reads are plotted for the intergenic region between moron CDS and downstream hypothetical ORF (indicated by red box above) for prophages encoding convergent sRNAs. Prophage designation and strand encoding PR’ are given in brackets. Peaks that have been assigned to predicted sRNA are indicated.
(D) Alignment of EcOnc01–3. Underlined sequence in EcOnc01 corresponds to the GcvB targeting consensus. The black triangle indicates the shortest alternate 5′ triphosphate end detected by 5′RLM-RACE in EcOnc03.