Figure 5.
The Shiga Toxin 2 Locus Encodes an Anti-sRNA that Enhances Expression of the Heme Oxygenase ChuS
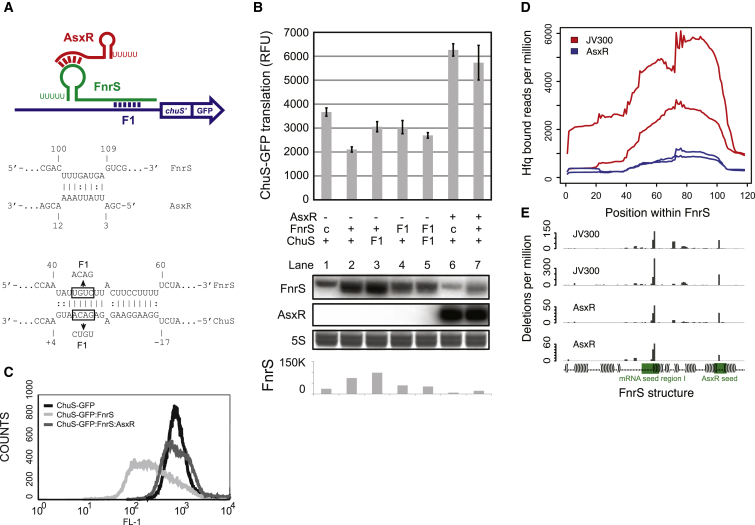
(A) (Top) Graphical representation of interactions between AsxR, FnrS, and the chuS mRNA. F1 indicates the positions of the complementary mutation. (Bottom) Predicted base paring (IntalRNA software) between AsxR and FnrS, and FnrS and the chuS transcript. Boxes and arrows indicate sequence changes that were introduced into F1 mutants.
(B) (Upper panel) Fluorescence of the 3′ chuA→5′ chuS chuS-GFP translational fusion was monitored in the presence of FnrS, AsxR, and appropriate point mutants (indicated below bar chart; basal levels of chromosomal FnrS are indicated by “c”). (Lower panel) Northern blot analysis of FnrS and AsxR (indicated). SYBR-green-stained 5S rRNA (5S) is included as a loading control. (Bottom) Quantification of FnrS northern blots by densitometry. Error bars indicate SEM.
(C) Flow cytometry quantification of fluorescence from cells expressing chuS-GFP alone, with FnrS, or with both FnrS and AsxR.
(D) AsxR reduces Hfq-bound FnrS. The chuS-GFP fusion and FnrS were constitutively expressed in E. coli MG1655 hfq-HTF with AsxR (blue) or the control plasmid pJV300 (red) and CRAC performed on these strains. Replicate data sets are plotted as reads per million across FnrS.
(E) Hfq binds to both seed and 3′ loop regions of FnrS. Deletions per million Hfq-bound reads are plotted relative to secondary structure of FnrS. Major deletion sites are located within the mRNA seed region I (green) and the AsxR seed region (green) within the terminator loop. See also Figure S3.