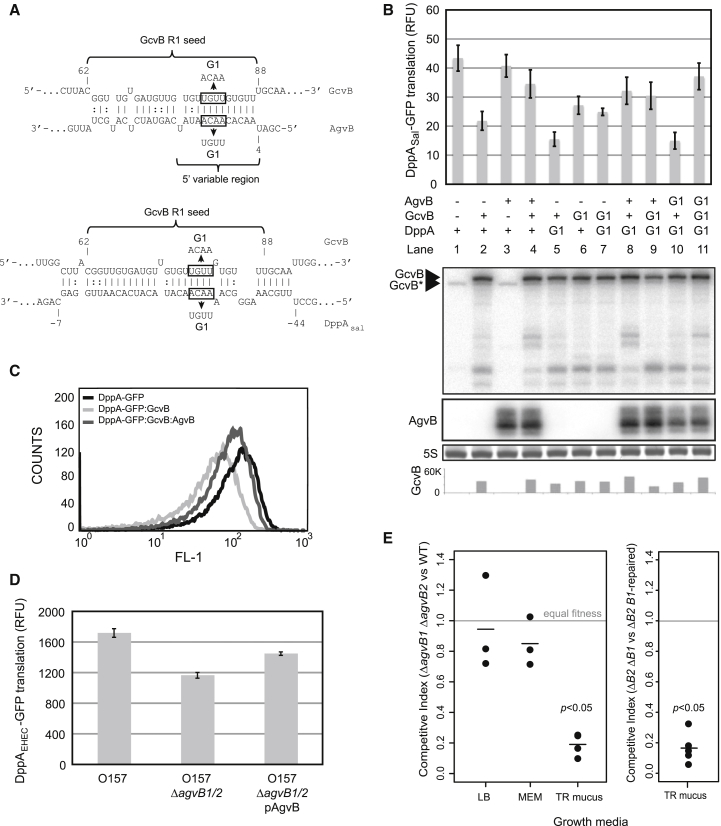
Figure 6.
EcOnc01 (AgvB) Acts as an “Anti-sRNA” to Inhibit GcvB Repression
(A) Interactions between GcvB and AgvB (top) and GcvB and DppASal (bottom) were predicted using IntaRNA software. The R1 seed sequence of GcvB is indicated in braces, and sequences that were introduced into G1 mutants are indicated within boxes.
(B) Fluorescence of DppAsal-GFP was used to monitor GcvB activity in the presence of AgvB, GcvB, and G1 mutants. Genotypes for each reading are indicated below. (Below: GcvB and AgvB) Northern analysis of GcvB and AgvB, respectively. GcvB∗ indicates the endogenous copy of GcvB, which carries an 8 nt deletion in the R1 seed region. SYBR-green-stained 5S rRNA (5S) is shown as a loading control for GcvB and AgvB northern blots. The bottom panel shows quantification of the exogenous copy of GcvB by densitometry. Error bars indicate SEM.
(C) Flow cytometry quantification of fluorescence from individual cells expressing DppAsal-GFP alone, with GcvB, or with both GcvB and AgvB.
(D) Fluorescence of DppAEHEC-GFP was used to monitor translation efficiency of DppA in E. coli O157:H7, ΔagvB1 ΔagvB2, and the complemented strain ΔagvB1 ΔagvB2 pZE12::EcOnc01 (pAgvB).
(E) The left-hand panel shows the competitive indices of E. coli O157 ΔagvB1 ΔagvB2 against the parent stain (Sakai) grown in LB media (n = 3), MEM-HEPES media (MEM, n = 3), and terminal rectal mucus (TR mucus, n = 4). The right-hand panel shows the competitive indices for the double mutant E. coli O157 ΔagvB1 ΔagvB2 against the same strain complemented on the chromosome with agvB1 (TR mucus, n = 5). A competitve index of 1 indicates no fitness difference; <1 indicates a fitness disadvantage.