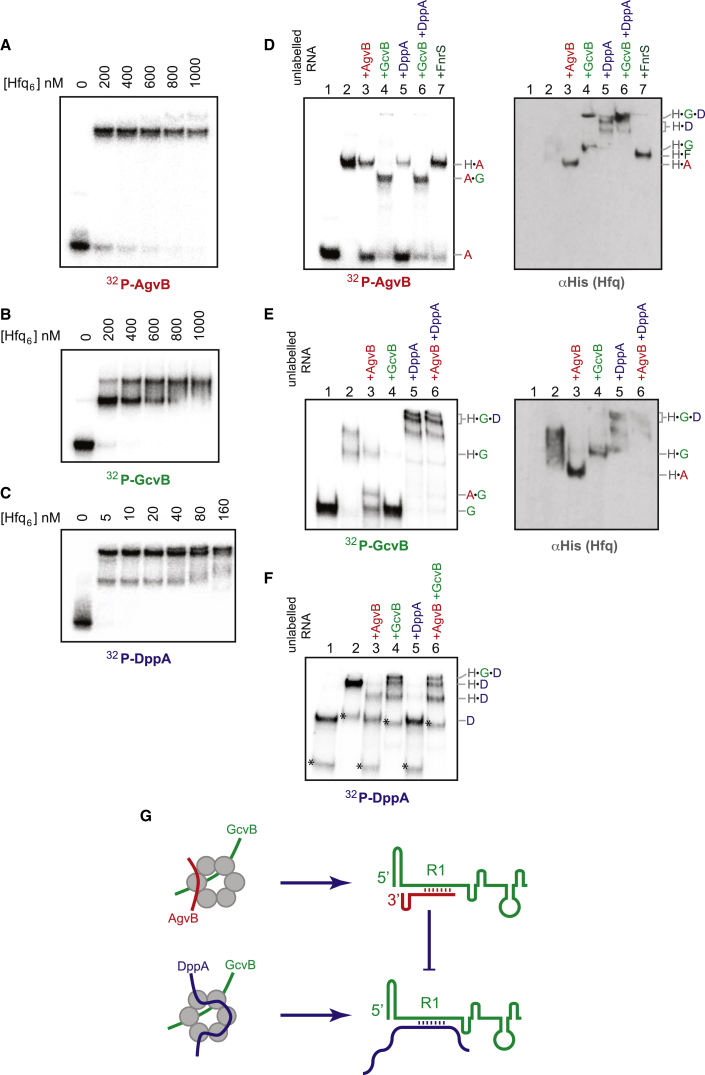
Figure 7.
EMSA Analysis in Hfq-AgvB Interactions
(A–C) Approximately 40 fmol of in-vitro-transcribed, radiolabeled AgvB (A), GcvB (B), or the 5′ 166 nt of DppASal (C) were incubated with increasing amounts of Hfq6 (indicated above).
(D–F) (Left panels) Competition assays with unlabelled RNAs. Radiolabelled AgvB (D), GcvB (E), or DppA (F), were incubated in the absence (lane 1) or presence of 500 nM Hfq6 (AgvB and GcvB) or 50 nM Hfq6 (DppA) (lanes 2–7). Hfq binding reactions were additionally incubated in the presence of a 50-fold excess of unlabelled competitor RNAs (indicated above gel, lanes 3–7). The composition of complexes is indicated on the right-hand side (H = Hfq, A = AgvB, G = GcvB, and D = DppA). For radiolabeled DppA (F), a shorter DppA RNA fragment copurified with the full-length product and is indicated by an asterisk.
(D and E) (Right panels) αHis western blot analysis of EMSA gels to monitor the presence of His6-tagged Hfq in gel-shifted complexes. Lanes are as in the left panels. In lanes E2 and E3, Hfq migrates as a smear, probably because it copurifies with heterogenous RNA species (Sittka et al., 2008), which are displaced in the presence of higher added concentrations of RNAs. In Figure 7F, the low Hfq concentration (50 nM) was not detectable by western analysis in DppA EMSA gels.
(G) Model for interaction of AgvB with Hfq, GcvB, and DppA. AgvB binds the distal face of Hfq (see also Figure S4) and forms a duplex with the R1 region of GcvB. Occlusion of the R1 region of GcvB prevents interactions between GcvB and the mRNA DppA. AgvB may also displace DppA from Hfq, although this interaction would be expected to be much more transient than inhibition through occlusion of GcvB R1. In the absence of AgvB, Hfq facilitates duplex formation between DppA and GcvB, repressing translation of DppA.