Figure 3.
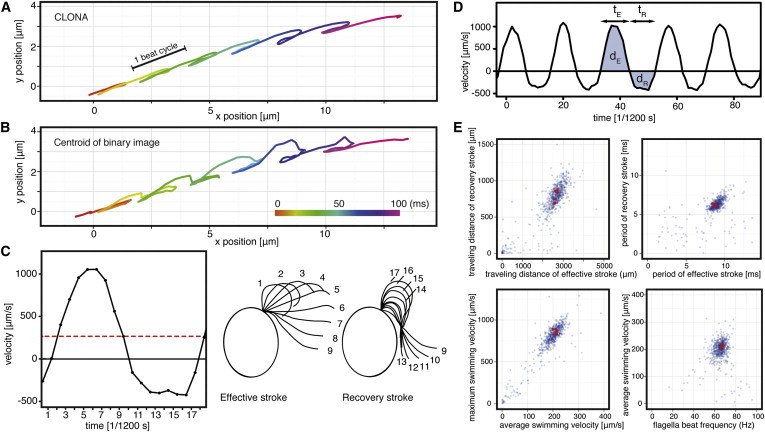
Application of the CLONA method to high-speed video images of wild-type Chlamydomonas cells (see Movie S1 in the Supporting Material). Swimming paths of a typical wild-type Chlamydomonas cell determined by (A) the CLONA methods and (B) conventional binary-centroid method. The position in each time frame is indicated by a dot and the frame interval is 1/1200 s ≈ 0.83 ms. The smooth curve indicates the better accuracy of the CLONA method in tracing the swimming path of Chlamydomonas cells. (C) The relation between the instantaneous swimming velocity parallel to the swimming direction and the flagellar beat stage obtained from Movie S2. The curve was obtained from a single beat without applying any filters. The mean swimming velocity is indicated by the red dotted line. (D) A typical plot of instantaneous swimming velocity. The following swimming parameters obtained from each beat cycle are shown: period of the effective and recovery strokes (tE and tR, respectively); travel distance of each stroke (dE and dR, respectively). (E) Scatter plots of wild-type beat cycles. Each point represents one beat cycle in each two-dimensional space. (Red points) Beat cycles of a single typical wild-type cell.
