Figure 1.
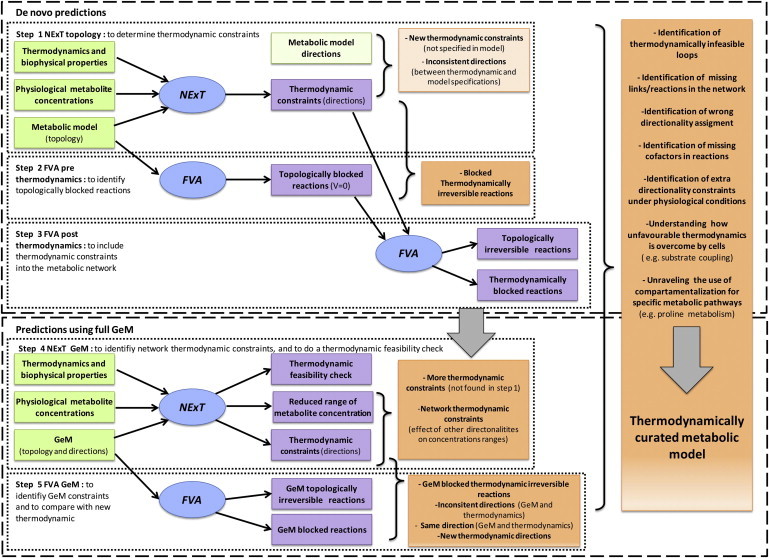
Methodology workflow. A five-steps process is used to generate a thermodynamically curated metabolic model. The first step (NExT topology) identifies thermodynamic constraints, which are compared with the model specifications. Step 2 (FVA prethermodynamics) identifies blocked reactions caused solely by network topology; together with the results from step 1, blocked thermodynamically irreversible reactions are thus identified. Step 3 (FVA post thermodynamics) identifies the reduction of the solution space due to thermodynamic constraints. Step 4 (NExT GeM) identifies new thermodynamic constraints that are a consequence of network-specified directionality constraints; the metabolite concentration ranges are also thermodynamically constrained. Step 5 (FVA GeM) identifies the solution space of the model using network-specified directionality constraints only, which are compared with results of step 4 to identify the new thermodynamic constraints that will reduce the flux solution space. Inputs to the process are in green boxes, outputs in purple boxes, and information inferred in orange boxes; simulations are in blue ovals.
