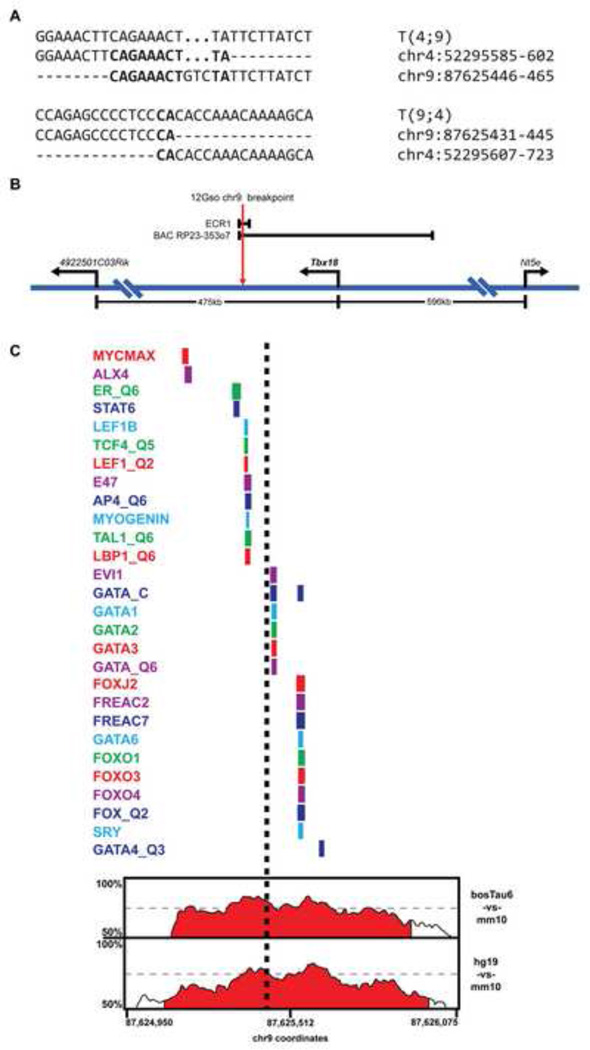
Fig. 3.
The 12Gso breakpoint. (A) Sequence of the 12Gso breakpoints on Mmu4;9 and Mmu9;4. Bold text indicates sequence homology between the two chromosomes. (B) Map of the Mmu9 Tbx18 locus (not drawn to scale). The 12Gso breakpoint is located 78 kbp downstream of the 3’ end of Tbx18. The enhancer ECR1 spans the 12Gso breakpoint and contains a highly conserved cluster of elements. The BAC RP23-353o7, which spans 210 kbp of the Tbx18 locus was reported to contain a somite and ureter enhancer, and extends just past the 12Gso Mmu9 breakpoint. (C) A map of mammalian conservation and conserved transcription factor binding motifs surrounding the 12Gso breakpoint within ECR1. At the bottom of the figure is a percent identity plot (PIP) from the ECR browser (Ovcharenko et al., 2004) showing sequence similarity across a ~1 kbp region surrounding the 12Gso breakpoint within the ECR1 sequence. Percent identity (between 50–100% in 100 bp windows) is traced over the region to identify regions with significant sequence homology between the mouse sequence and regions in the human (hg19), bovine (bostTau6), and opossum (monDom5) genomes. The conserved regions are filled with red by default in the ECR browser if they include sequence identity above 70% in 100 bp windows. Above the PIP plot is a map of the locations of transcription factor binding motifs, identities of which are listed at left, that are conserved in all four species. The approximate location of the 12Gso breakpoint is shown as a black dashed line.