Figure 4. Changes in CBF with normalization of CO2 after hypocapnia are similar under control conditions, hyperglycemia or ketosis.
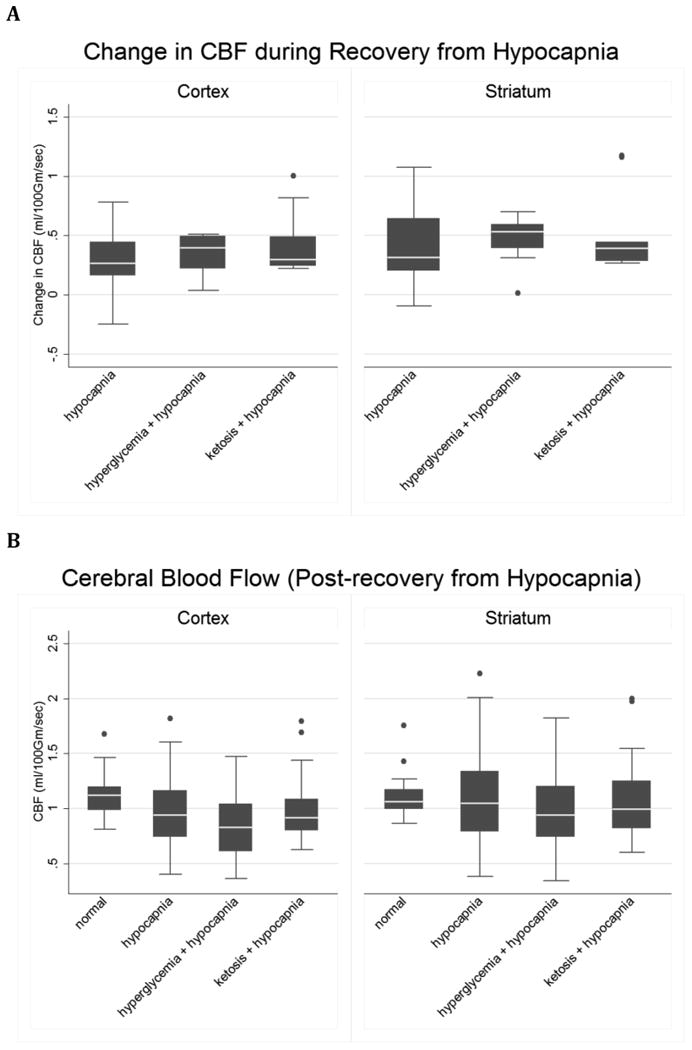
A. Control (n=14), hyperglycemia (n=12) and ketosis (n=9). The line in the middle of each box indicates the median (50% percentile), the top and bottom indicate the 75th / 25th percentiles. The whiskers (1.5 interquartile ranges) describe the maximum and minimum values. Individual values beyond the whiskers are possible outliers. Cortex: Oneway ANOVA omnibus F-test: F(2,32)=1.37, p= 0.27. Striatum: Omnibus F(2,32)=0.35, p= 0.70. B. CBF in the cortex and striatum in control (n=17), and in hypocapnic rats after CO2 normalization: hypocapnia only (H), hyperglycemia plus hypocapnia (G) and ketosis plus hypocapnia (K). Pairwise contrasts in means (95% CI) and p-values were estimated using Tukey-Kramer multiple comparisons procedure to control the familywise Type-1 error rate, with statistical significance declared for simultaneous p < 0.05. Cortex: Omnibus F(3, 48) = 1.61, p = 0.20. Model R-square=0.09; root mean square error=0.30. K vs. N = 0.16 ( -0.17, 0.48 ), p=0.58; H vs. N = 0.09 ( -0.20, 0.37 ), p=0.84; G vs. N = -0.10 ( -0.40, 0.19 ), p=0.79 ; K vs. H = 0.07 ( -0.27, 0.40 ), p=0.96 ; K vs. G = 0.26 ( -0.09, 0.61 ), p=0.20 ; H vs. G = 0.19 ( -0.12, 0.50 ), p=0.36 . Striatum: Omnibus F(3, 48) = 3.15, p=0.03. Model R-square=0.16; root mean square error=0.34. H vs. N=0.34 ( 0.01, 0.67 ), p=0.04; K vs. N=0.31 ( -0.07, 0.69 ), p=0.14; G vs. N=0.12 ( -0.23, 0.46 ), p=0.80; H vs. K=0.03 ( -0.36, 0.43 ), p=0.995; H vs. G = 0.23 ( -0.13, 0.59 ), p=0.35; K vs. G = 0.19 ( -0.21, 0.59), p=0.59.
