Fig. 1. Vps20 binding to membranes.
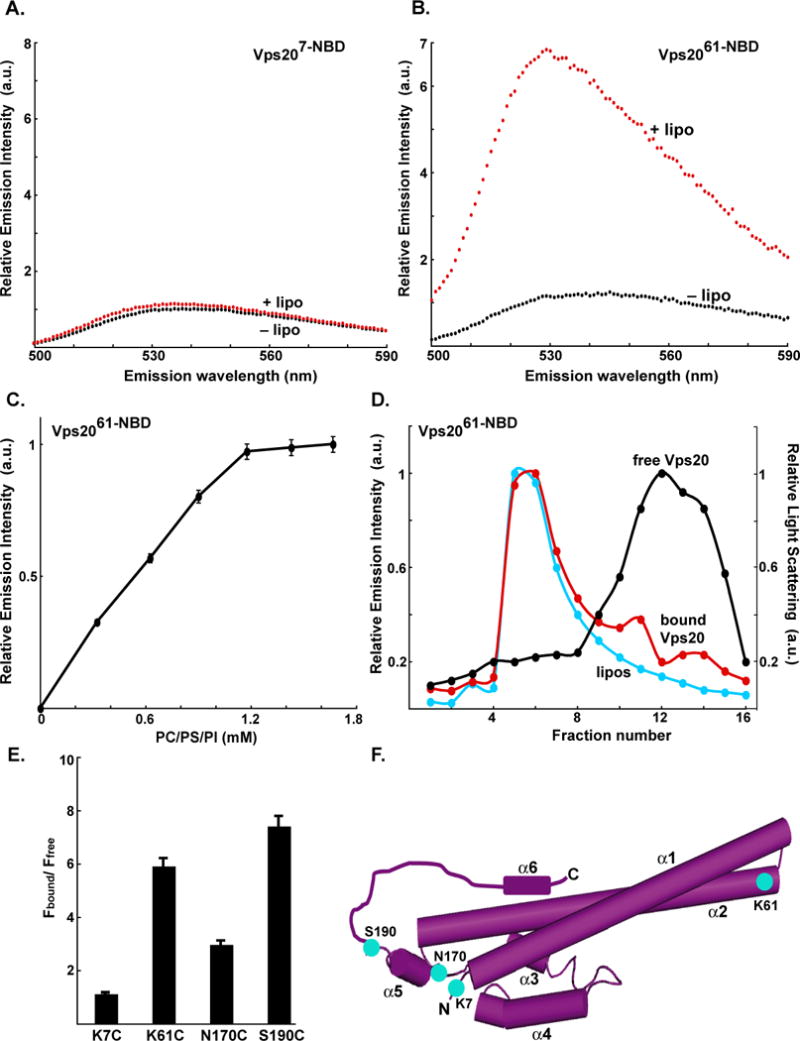
Fluorescence emission spectra of 750 nM Vps207-NBD (A) and 1 μM Vps2061-NBD (B) before (black) and after (red) addition of excess (1.6 mM; see Fig. 1C) PC/PS/PI. (C) Titration of 1 μM Vps2061-NBD with liposomes reveals that fluorescence-detected (λem = 530 nm) binding is complete after addition of 1.2 mM PC/PS/PI. (D) Sepharose CL-2B chromatography of 800 nM Vps2061-NBD that had been preincubated with either 1.5 mM PC/PS/PI (red) or no liposomes (black). Protein was detected by NBD emission intensity and liposomes by light scattering (cyan). The elution profile of liposomes that were not exposed to protein was identical to the profile shown in this and later figures (data not shown). (E) The ratios of NBD emission intensities at 530 nm are shown for each derivative with (Fbound) and without (Ffree) membranes. (F) Cartoon representation of the water-soluble hVps24 monomer. Based on the sequence similarities of the ESCRT-III proteins, Vps20, Snf7 and Vps2 are believed to share structural features with the solved crystal structure of hVps24. Helices α1–5 and the N- and C-termini of the protein are labeled. Residues that were mutated to cysteine for labeling with NBD are depicted in cyan and labeled accordingly. Helix α6 as well as the α5–α6 loop have been manually sketched.
