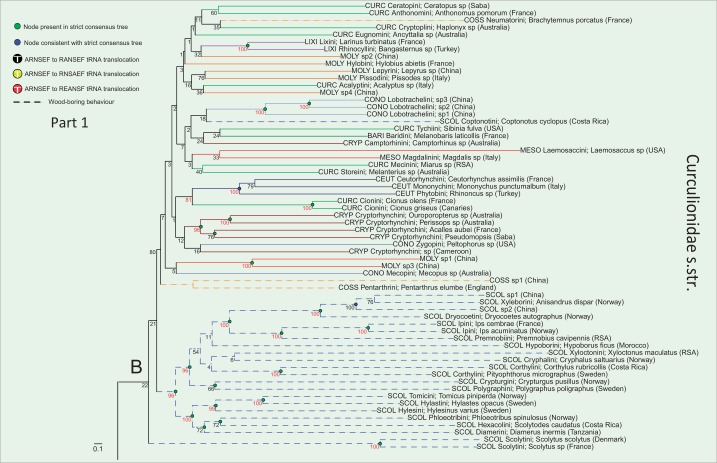
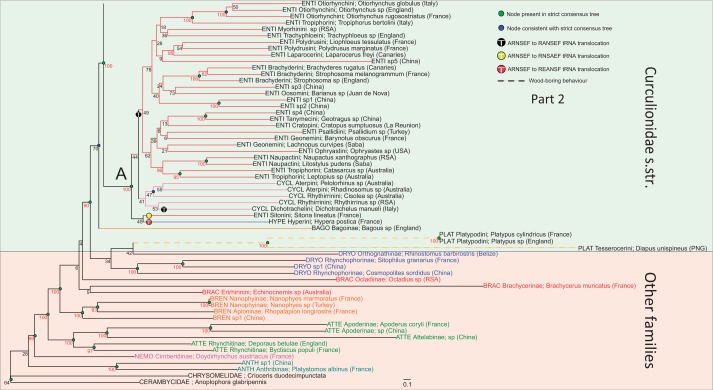
Fig. 5.
(Parts 1 and 2) ML tree resulting from the analysis of the “all genes” data set partitioned according to the six PartitionFinder partitions (see table 1). Within Curculionidae s. str. (sensu Bouchard et al. 2011) branches are colored according to subfamily. Other curculionoid families have their name labels colored by family. Numbers adjacent to nodes are RAxML rapid bootstrap scores, with values more than 80% highlighted in red. The three principal wood-boring subfamilies are represented by dashed branches and the nodes labeled A and B indicate the two large divisions within Curculionidae referred to in the text. Nodes indicated in green correspond to nodes present in the strict consensus tree and nodes indicated in blue are consistent with it. The positions of the three tRNA rearrangements are indicated. Scale bar represents substitution rate. Family and subfamily codes precede taxa names as follows: Anthribidae (ANTH), Attelabidae (ATTE), Brachyceridae (BRAC), Brentidae (BREN), Dryophthoridae (DRYO), Nemonychidae (NEMO), Bagoinae (BAGO), Baridinae (BARI), Ceutorhynchinae (CEUT), Conoderinae (CONO), Cossoninae (COSS), Cryptorhynchinae (CRYP), Curculioninae (CURC), Lixinae (LIXI), Mesoptillinae (MESO), Molytinae (MOLY), Platypodinae (PLAT), and Scolytinae (SCOL).