Abstract
Although the structure and function of the AhR are conserved, emerging evidence suggests that downstream effects are species-specific. In this study, rat hepatic gene expression data from the DrugMatrix database (National Toxicology Program) were compared to mouse hepatic whole-genome gene expression data following treatment with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). For the DrugMatrix study, male Sprague–Dawley rats were gavaged daily with 20 μg/kg TCDD for 1, 3 and 5 days, while female C57BL/6 ovariectomized mice were examined 1, 3 and 7 days after a single oral gavage of 30 μg/kg TCDD. A total of 649 rat and 1386 mouse genes (|fold change|≥1.5, P1(t)≥0.99) were differentially expressed following treatment. HomoloGene identified 11,708 orthologs represented across the rat Affymetrix 230 2.0 GeneChip (12,310 total orthologs), and the mouse 4×44K v.1 Agilent oligonucleotide array (17,578 total orthologs). Comparative analysis found 563 and 922 orthologs differentially expressed in response to TCDD in the rat and mouse, respectively, with 70 responses associated with immune function and lipid metabolism in common to both. Moreover, QRTPCR analysis of Ceacam1, showed divergent expression (induced in rat; repressed in mouse) functionally consistent with TCDD-elicited hepatic steatosis in the mouse but not the rat. Functional analysis identified orthologs involved in nucleotide binding and acetyltransferase activity in rat, while mouse-specific responses were associated with steroid, phospholipid, fatty acid, and carbohydrate metabolism. These results provide further evidence that TCDD elicits species-specific regulation of distinct gene networks, and outlines considerations for future comparisons of publicly available microarray datasets.
Keywords: Mouse, Rat, Cross-species, TCDD, Microarray
Introduction
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is associated with species-specific responses that are mostly mediated by the aryl hydro-carbon receptor (AhR) (Denison and Heath-Pagliuso, 1998; Denison et al., 2011; Gonzalez and Fernandez-Salguero, 1998). TCDD binding to the AhR leads to the dissociation of chaperone proteins HSP90, p23, and AIP (also known as ARA9 or XAP2), heterodimerization with the AhR nuclear translocator (ARNT), and translocation of the complex to the nucleus (Hankinson, 1995). Liganded AhR–ARNT complexes bind to dioxin response elements (DREs) consisting of a core sequence 5′-GCGTG-3′ in the promoter region of target genes, altering their expression (Hankinson, 1995), although recent studies indicate that non-canonical mechanisms independent of DREs serve an underappreciated role in the effects elicited by TCDD and related compounds (Beischlag et al., 2008; Denison et al., 2011; Dere et al., 2011c; Huang and Elferink, 2012).
Despite similarities in structural organization, amino acid sequence, and mode of action, as well as the conserved specificity of AhR binding to DREs (Bank et al., 1992; Denison et al., 1986, 2011; Hahn et al., 1997; Okey et al., 1994), a number of species-specific toxicogenomic, histopathological, and physiological responses are elicited by TCDD and related compounds (Black et al., 2012; Boutros et al., 2008; Boverhof et al., 2006; Carlson et al., 2009; Dere et al., 2011b; Forgacs et al., 2012, submitted for publication; Gonzalez and Shah, 2008; Maglich et al., 2002; Poland and Knutson, 1982; Silkworth et al., 2005). For example, acute high dose TCDD exposure induces hepatic steatosis in the mouse, with minimal hepatic fat accumulation in rats, while hepatocyte hyper-trophy is only reported in rat (Boverhof et al., 2006). Species-specific differences in choline metabolism (Forgacs et al., 2012) and serum clinical chemistry responses including alterations in serum alanine aminotransferase, cholesterol, free fatty acids, and triglycerides have also been reported (Boverhof et al., 2006; Forgacs et al., 2012).
Species-specific responses confound the extrapolation of rodent data for use in human risk assessment (Olson et al., 2000; Smith, 2001). To further investigate the species-specific hepatic effects of TCDD, comparative gene expression profiles have been examined (Black et al., 2012; Boutros et al., 2008; Boverhof et al., 2006; Carlson et al., 2009; Dere et al., 2011b). In vivo genome-wide analysis showed that <20% of TCDD-responsive orthologs were conserved between rats and mice. Many of these differences were also reflected in species-specific hepatic metabolomic profiles (Forgacs et al., 2012). Similarly, >70% of the TCDD-elicited differentially expressed orthologs in rat H4IIE, mouse Hepa1c1c7, and human HepG2 hepatoma or HL1-1 liver stem-cell like cell lines were species-specific (Dere et al., 2011b; Kim et al., 2009). However, these studies were limited by the number of comparable rat orthologs represented on the microarrays (3087 and 8125 in the Boverhof et al., 2006 and Boutros et al., 2008 studies, respectively).
Recently, the DrugMatrix database which contains genome-wide rat microarray datasets for over 600 chemicals was made publicly available by the National Toxicology Program (NTP). In this study, the more comprehensive DrugMatrix rat dataset was used to expand our previous comparison of TCDD-elicited genome-wide in vivo rat and mouse hepatic gene expression using the same normalization and analysis methods. This cross-platform comparison involved 11,708 orthologs represented in both datasets, thus addressing the coverage limitations of previous in vivo studies. We show that >85% of TCDD-responsive orthologs are species-specific providing further evidence of TCDD elicited species-specific hepatic gene expression (Black et al., 2012; Boutros et al., 2008; Boverhof et al., 2006; Budinsky et al., 2010; Carlson et al., 2009; Dere et al., 2011b; Forgacs et al., submitted for publication; Silkworth et al., 2005) and species-specific ortholog differential expression associated with lipid, glycerolipid and carbohydrate metabolism, supporting divergent in vivo metabolic responses to TCDD. Moreover, we also outline several considerations for future comparisons to publicly available datasets such as those found in the DrugMatrix database.
Materials and methods
Microarray datasets
Rat microarray raw data were retrieved from the DrugMatrix database (https://ntp.niehs.nih.gov/drugmatrix; provided by Dr. S. Auerbach, National Toxicology Program). Gene expression studies used male Sprague–Dawley rats (6–8 weeks old) orally gavaged daily with carboxymethyl cellulose (CMC; vehicle control) or 20 μg/kg body weight TCDD in CMC, and euthanized at 1, 3 and 5 days post-dose (Ganter et al., 2005). Hepatic gene expression was assessed using the Affymetrix rat genome GeneChip 230 2.0 arrays (Santa Clara, CA).
The mouse microarray dataset used 4×44K Agilent oligonucleotide arrays (version 1) (Dere et al., 2011c) and hepatic samples from a published study (Boverhof et al., 2005). Briefly, ovariectomized (ovx) immature female C57BL/6 mice were gavaged once with sesame oil vehicle or 30 μg/kg TCDD in sesame oil on postnatal day 28. Mice were euthanized at 2, 4, 8, 12, 18, 24, 72 and 168 h, and total hepatic RNA was extracted. For the purpose of this study, only 24, 72 and 168 h (1, 3 and 7 days) were retrieved from dbZach (Burgoon et al., 2006) and re-analyzed for comparison.
Microarray data analysis, functional annotation and clustering
Rat raw microarray signal intensity values were generated using the MAS5 algorithm implemented in the Affymetrix Expression Console (http://affymetrix.com). Rat and mouse raw microarray signal intensity values were then normalized in-house using our semiparametric approach (Eckel et al., 2005) in SAS version 9.1 (Cary, NC). Subsequently, the normalized data from both studies were analyzed using the same approach to facilitate comparisons. An empirical Bayes method was used to calculate posterior probability P1(t) values on a per-gene and -time point basis (Eckel et al., 2004) using R v1.8.1 (http://www.r-project.org). Genes were considered differentially expressed using |fold change|≥1.5 and statistical P1(t)≥0.99 cut-offs. Orthologs were identified using HomoloGene identifiers (build 66; http://www.ncbi.nlm.nih.gov/HomoloGene/).
Functional annotation and categorization of orthologs were performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov) (Dennis et al., 2003). Functionally clustered orthologs were filtered for gene ontology biological processes (BP) and molecular functions (MF). Scores≥1.3 (equivalent to a -log scale geometric mean p-value of 0.05) were considered significantly enriched. Transcription factor (TF) analysis (Ingenuity Pathway Analysis, Ingenuity Systems, Redwood City, CA; www.ingenuity.com) was used to predict active and inhibited transcriptional regulators (TRs). TF analysis uses a proprietary algorithm to analyze downstream gene expression to estimate the likelihood of a TR being (or having been) activated or inhibited based on a calculated Z-score (IPA, 2011).
Quantitative real-time PCR (QRTPCR) of Ceacam1
Hepatic RNA isolation, cDNA plate preparation, and QRTPCR were performed as previously described (Boverhof et al., 2005; Kopec et al., 2011). Rat hepatic samples from the DrugMatrix database were not available for QRTPCR. RNA extracted from frozen liver samples of female Sprague–Dawley rats (Boverhof et al., 2006) and female C57BL/6 ovx mice (Kopec et al., 2008) orally gavaged once with TCDD in sesame oil at 10 μg/kg and 30 μg/kg, respectively, for 1, 3, and 7 days were used. Reactions were performed in 96-well plates (Applied Biosystems, Foster City, CA) containing 1.0 μl cDNA template, 0.1 μM gene-specific forward and reverse primers, 3 mM MgCl2, 1.0 mM dNTPs, 0.025 IU Amplitaq Gold and 1 × SYBR Green PCR buffer (Applied Biosystems) in a final volume of 30 μl and amplified using an Applied Biosystems PRISM 7000 sequence detection system. Rat and mouse Ceacam1 cDNAs were quantified using a standard curve and standardized to the geometric mean of housekeeping genes ActB, Hprt, and Gapd or Rpl13 (Vandesompele et al., 2002). Primer sequences for housekeeping genes were previously described (Boverhof et al., 2006) and Ceacam1 primer sequences are listed in Supplementary Table S1. QRTPCR data were analyzed by analysis of variance (ANOVA) followed by Tukey's post hoc test using SAS 9.1.
Genomic AhR ChIP–chip and DRE enrichment analysis
AhR enrichment of genomic regions was determined using whole-genome ChIP–chip data (Dere et al., 2011c). Putative DRE (pDRE) distributions within rat and mouse genes were identified using a position weight matrix (PWM) (Dere et al., 2011a). Briefly, mouse ChIP–chip analysis was performed using an Affymetrix GeneChip mouse 2.0R tiling array (Dere et al., 2011c). A 1% false discovery rate (FDR≤0.01) statistical cut-off was determined using the default moving average (MA) approach in TileMap (Dere et al., 2011c). pDREs were determined using a PWM based on bona fide DRE sequences comprising the DRE core sequence (5′-GCGTG-3′) and the adjacent 7 bp upstream and downstream flanking regions. Although a matrix similarity score (MSS) of 0.8473 is suggested as the threshold for identifying pDREs (Dere et al., 2011a), DREs with MSS≥0.8 were considered candidates as pDREs for AhR complex binding. pDREs and AhR regions of enrichment within the Ceacam1 genomic region were aligned using custom tracks in the University of California—Santa Cruz Genome Browser (Fujita et al., 2011) for rat (build rn4) and mouse (build mm9) (Dere et al., 2011c).
Results
Re-analysis and re-annotation of rat Affymetrix dataset
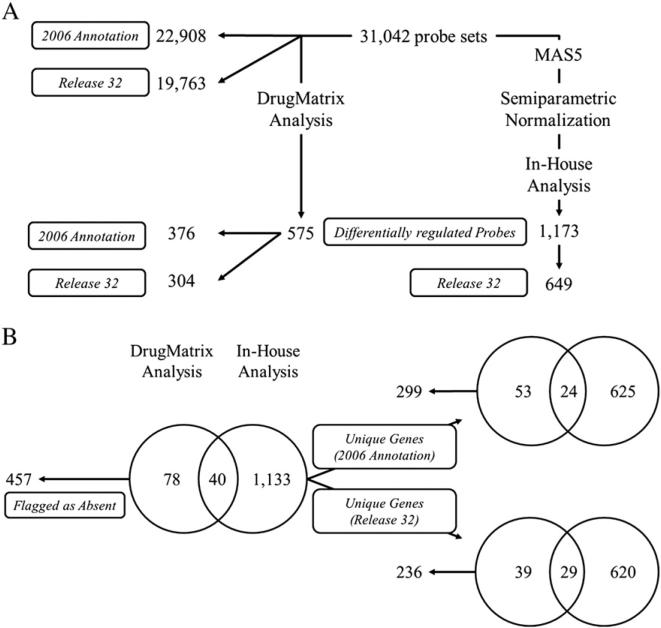
Rat Affymetrix data was last annotated in 2006 and normalized using a nonlinear normalization procedure and empirical Bayes analysis approach developed by Iconix, the DrugMatrix developers (Ganter et al., 2005; NIEHS/NTP, 2011). In order to facilitate a more equitable comparison to our mouse gene expression dataset (Dere et al., 2011c), raw probe signals were extracted from the Affymetrix GeneChip 230 2.0 array CEL files, and probe set calls determined using the MAS5 algorithm (http://affymetrix.com) These data were then re-analyzed using our in-house semiparametric normalization and empirical Bayes statistical methods (Eckel et al., 2004, 2005). Re-annotation using release 32 (August 2011; http://www.affymetrix.com/support/help/releasedocs/netaffx_release_32.affx) also decreased the number of genes represented by the 31,042 probe sets of the Affymetrix GeneChip 230 2.0 from 22,908 to 19,763 Gene IDs. Only ~71% of the 31,042 probe sets have the same annotation in release 32 compared to the 2006 annotation.
Using nonlinear normalization and an empirical Bayes analysis, Iconix originally identified 575 differentially expressed probe sets that represent 304 genes based on release 32 annotation (or 376 genes using 2006 annotation) (Ganter et al., 2005; NIEHS/NTP, 2011). However, different normalization and analysis procedures can significantly influence the identification of differentially expressed genes (Guo et al., 2006; Shi et al., 2006). Using the MAS5 algorithm to determine rat Affymetrix calls for each probe set, the calls were then subjected to our semiparametric normalization and empirical Bayes statistical analysis, 1173 differentially regulated probe sets (|fold change|≥1.5 and P1(t)≥0.99) representing 649 unique genes (Fig. 1A) were identified. This provided a differentially expressed gene list for the rat Affymetrix dataset that could be compared to the mouse Agilent datasets identified using the same normalization and data analysis approaches, thus minimizing confounding variables introduced by different data analysis approaches. Comparison of the Iconix analysis published within the DrugMatrix database to our in-house analysis identified 40 overlapping differentially expressed probe sets within the rat Affymetrix dataset (Fig. 1B). Using 2006 annotation, the 40 commonly regulated probe sets represent 24 unique genes but only 29 differentially expressed unique genes based on release 32 annotation (Fig. 1B). Further examination identified 457 probe sets that were excluded from the comparison due to absent detection calls by MAS5 which likely contributed to the poor overlap between the two analysis methods.
Fig. 1.
Comparison of the rat Affymetrix GeneChip 230 2.0 probe set annotation and differentially regulated probes. (A) Differentially regulated probes of the previously analyzed DrugMatrix dataset using an empirical Bayes analysis for estimation of variance and p-values (p≤0.05) or following re-annotation (Affymetrix annotation release 32; http://www.affymetrix.com/support/help/releasedocs/netaffx_release_32.affx), semiparametric normalization, and empirical Bayes estimation of posterior probabilities (in-house analysis) examined for changes in annotation using the 2006 annotation found in the online database or following re-annotation by Affymetrix release 32. (B) Overlap of differentially regulated probe sets and unique genes identified by the DrugMatrix analysis or in-house analysis using the 2006 annotation or Affymetrix annotation release 32. Probes and unique genes flagged as absent were determined using the MAS5 algorithm implemented in the Affymetrix Expression Console (http://affymetrix.com).
Comparative analysis of rat and mouse microarray datasets
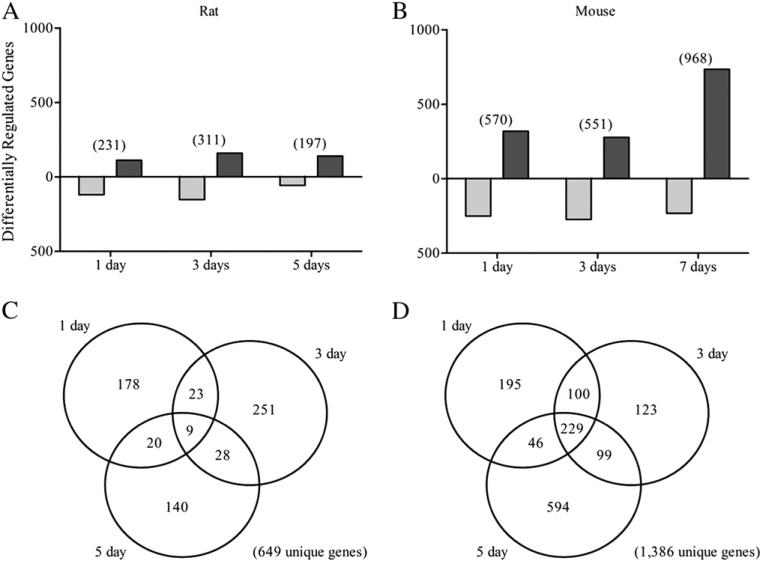
A total of 13,874 and 21,307 unique genes are represented on the rat Affymetrix GeneChip 230 2.0 and mouse Agilent 4×44K oligonucleotide array, respectively. Semiparametric normalization (Eckel et al., 2005) and empirical Bayes analysis (Eckel et al., 2004) identified 649 differentially expressed rat genes (|fold change|≥1.5 and P1(t)≥0.99) across days 1, 3 and 5 based on release 32 annotation while 1386 mouse genes are differentially expressed across 1, 3 and 7 days (Fig. 2) using mm9 genome build annotation. Rat differential expression showed distinct temporal patterns with only ~1% (9 genes) active across all three time points compared to the >16% of mouse genes active at 1, 3 and 7 days. Rat differential expression ranged from the ~14-fold induction for Nr1d1 to ~6.7-fold repression for cxcl1, both at 5 days. Cyp1a1 was not reported to be induced in the rat study. This was not attributed to signal variability or probe design but may be due to the use of carboxymethyl cellulose (CMC) as the vehicle resulting in an underestimation of administered TCDD dose (Dr. S. Auerbach, personal communication). However, similar response of several other genes including Cpt1a, Mt1a, Pdk2, and Srebf1 to previous rat gene expression studies (Boverhof et al., 2006) and observation of minimal lipid accumulation, inflammation, and necrosis at a lower dose (0.7 μg TCDD/kg/day in CMC vehicle) as early as 5 days (https://ntp.niehs.nih.gov/drugmatrix) suggest that satisfactory TCDD exposure did in fact occur. Mouse temporal differential expression exhibited almost twice as many differentially expressed genes (570, 551, 968 at 1, 3 and 7 days, respectively) with Cyp1a1 (~206-fold) and Serpina7 (~8-fold) representing the most induced and repressed genes, respectively (Dere et al., 2011a,b,c).
Fig. 2.
Hepatic differential gene expression in the rat and mouse. Gene annotation for the rat dataset was first updated using Affymetrix release 32 annotation (http://www.affymetrix.com/support/help/releasedocs/netaffx_release_32.affx). Updated rat Affymetrix GeneChip 230 2.0 and mouse 4×44K Agilent oligonucleotide array (version 1) raw datasets were normalized by our in-house semiparametric normalization (Eckel et al., 2005), and empirical Bayes analysis methods (Eckel et al., 2004). Temporal distribution of differentially expressed genes (|fold change|≥1.5 and P1(t)≥0.99) in (A) rat and (B) mouse per time point (repressed — light gray; induced — dark gray). The total number of responsive genes at each time point is indicated in parentheses. Venn diagrams compare differentially regulated genes at each time point for (C) rat and (D) mouse.
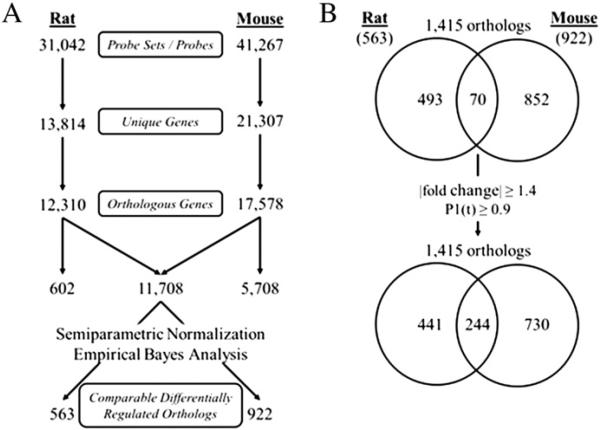
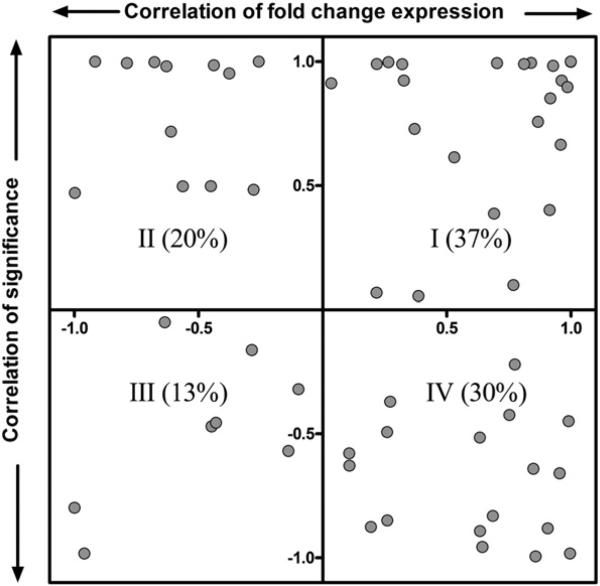
HomoloGene (build 66; http://www.ncbi.nlm.nih.gov/HomoloGene/) identified 11,708 orthologs represented on both platforms, of which 563 and 922 were differentially expressed (|fold-change|≥1.5, P1(t)≥0.99) in the rat and mouse, respectively (Fig. 3). Only 70 orthologs were differentially expressed in both species (Fig. 3; Supplementary Table S2). Although relaxation of the filtering criteria to |fold-change|≥1.4, P1(t)≥0.9 increased the overlap to 244 differentially expressed orthologs, the results are consistent with TCDD eliciting species-specific gene expression profiles. Moreover, the correlation analysis of the 70 commonly regulated orthologs using fold change and P1(t) values found only ~37% exhibit comparable expression profiles (Fig. 4, Quadrant I).
Fig. 3.
Comparison of hepatic rat and mouse TCDD-elicited differential ortholog expression. (A) Comparison of unique genes, orthologs and TCDD-responsive orthologs in rat and mouse. Gene annotation for the rat Affymetrix dataset was first updated using Affymetrix release 32 annotation (http://www.affymetrix.com/support/help/releasedocs/netaffx_release_32.affx). Both the updated rat Affymetrix GeneChip 230 2.0 and the mouse 4×44K Agilent oligonucleotide array (version 1) raw datasets were normalized by a semiparametric method (Eckel et al., 2005), and statistically analyzed using an empirical Bayes method (Eckel et al., 2004). (B) Venn diagrams of 1415 comparable differentially regulated orthologs (563 rat and 922 mouse) representing species-specific and commonly regulated TCDD-responsive orthologs using stringent (|fold change|≥1.5, P1(t)≥0.99) and relaxed criteria (|fold change|≥1.4, P1(t)≥0.90).
Fig. 4.
Correlation analysis of commonly regulated TCDD-responsive genes (|fold change|≥1.5, P1(t)≥0.99). Gene annotation for the rat Affymetrix dataset was first updated using Affymetrix release 32 annotation (http://www.affymetrix.com/support/help/releasedocs/netaffx_release_32.affx). The updated rat Affymetrix GeneChip 230 2.0 and the mouse 4×44K Agilent oligonucleotide array (version 1) raw datasets were normalized by a semiparametric method (Eckel et al., 2005), and statistically analyzed using an empirical Bayes method (Eckel et al., 2004). Only 37% genes were positively correlated in terms of fold change and significance between rat and mouse genes (quadrant I).
Functional annotation of the 70 commonly regulated orthologs using DAVID identified over-represented functions associated with external stimuli, chemokine activity, cell proliferation, lipid catabolism and regulation of transcription. More specifically, lipid catabolism genes phospholipase A2 group XIIA (Pla2g12a; induced up to ~4.5-fold) and patatin-like phospholipase domain containing 3 (Pnpla3; repressed up to ~3.4-fold) were differentially expressed in both species. Lipoprotein lipase (Lpl) was the only lipid metabolism related ortholog to show divergent regulation (repressed ~1.5-fold in rat; induced ~3.0-fold in mouse). Other orthologs involved in lipid metabolism common to both species include sterol regulatory element binding factor 1 (Srebf1; repressed up to ~1.7-fold), fatty acid elongase 6 (Elovl6; repressed up to ~2.8-fold), and hydroxysteroid (17-β) dehydrogenase 2 (Hsd17b2; induced up to ~3.4-fold). While similar responses of Srebf1 are reported in female rats (Boverhof et al., 2006), responses of Elovl6 and Hsd17b2 may be sex-specific.
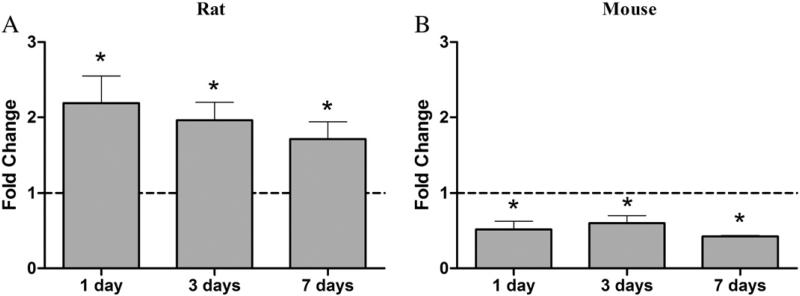
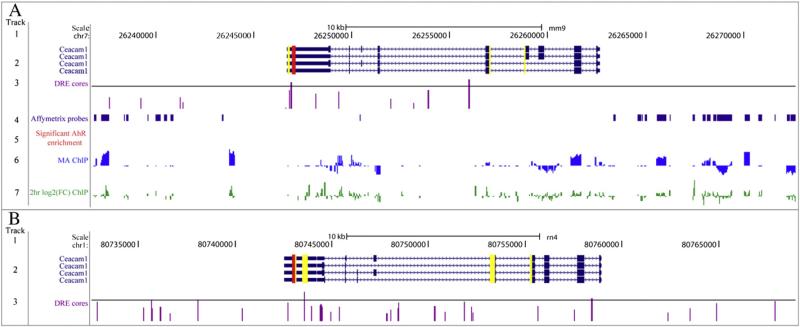
Ceacam1 is involved in insulin signaling and recycling (Najjar, 2002). In humans with fatty liver, Ceacam1 is repressed (Lee, 2011) while Ceacam1 null mice develop hyperinsulinemia and hepatic steatosis (DeAngelis et al., 2008). In contrast, Ceacam1 is induced by TCDD in rats (Fletcher et al., 2005), consistent with the lack of AhR-mediated hepatic steatosis (Boverhof et al., 2006). QRTPCR confirmed ~2-fold induction of Ceacam1 in the rat and ~2.4-fold repression in the mouse (Fig. 5). Several 19 bp DRE sequences were also identified within the 10 kb transcription start site upstream region of the rat Ceacam1 loci, three of which had an MSS>0.8 (Fig. 6, Supplementary Table S3).
Fig. 5.
Temporal QRTPCR analysis of hepatic Ceacam1 expression in (A) rat and (B) mouse liver. Bars represent gene expression relative to time-matched vehicle controls±standard error for at least 4 biological replicates. Data are scaled such that expression levels of vehicle controls are equal to one (dotted line). Asterisks (*) represent significant differences (p<0.05) compared to time-matched vehicle control determined by ANOVA followed by Tukey's post hoc test.
Fig. 6.
Examination of the rat and mouse Ceacam1 genomic regions. Computationally identified DRE core location and ChIP–chip AhR enriched regions have been previously reported (Dere et al., 2011a,c). Track 1 represents scale and chromosome position. Track 2 depicts the organization of the Ceacam1 genomic region including the TSS, exons (boxes), introns (arrows), and transcriptional variants. Yellow boxes indicate regions targeted by probes (Agilent or Affymetrix oligonucleotides), while red boxes represent the region targeted by QRTPCR primers used in this study. Track 3 summarizes the location of computationally identified DRE core sequences. The height of the purple lines provides a qualitative measure of the matrix similarity scores (MSS) for the 19 bp DRE sequence. The horizontal line indicating an MSS=0.8473 identifying 19 bp DRE sequences with putative function. Tracks 4–7 for mouse summarize in vivo hepatic ChIP–chip AhR regions of enrichment (Dere et al., 2011c). Track 4 illustrates probe tiling for the Affymetrix 2.0R mouse chip. Track 5 identifies regions significantly enriched (FDR≤0.01) for AhR enrichment. Tracks 6 and 7 are histograms representing the moving average (MA, track 6) and log2 fold enrichment (track 7) for regions of AhR enrichment.
Species-specific TCDD-responsive orthologs
Rat- and mouse-specific expression represents 88% and 92% of the total differentially expressed orthologs, respectively. Ratspecific responses were associated with antigen processing and presentation, nucleotide binding, and acetyltransferase activity categories. Over-represented mouse-specific functions included immune responses, steroid metabolism, fatty acid metabolism, glycerolipid metabolism, and xenobiotic metabolism (Table 1). Interestingly, rat-specific acetyltransferase activity was linked to chromatin remodeling and phospholipid metabolism. The latter includes 1-acyl-sn-glycerol-3-phosphate acyltransferase gamma (Agpat3) and lysophosphatidylcholine acyltransferase 1 (Lpcat1) involved in the production of phosphatidic acid and phosphatidylcholine from lysophosphatidic acid and lysophosphatidylcholine, respectively. Both orthologs were induced ~1.5-fold at day 5. Moreover, the phospholipid metabolism genes mitochondrial glycerol-3-phosphate acyltransferase (Gpam) and phospholipase A2 group XV (Pla2g15) were suppressed (~2-fold) and induced (~1.6-fold), respectively. In contrast, mouse specific induced genes included arachidonate 12-lipoxygenase (Alox12; ~1.6-fold) involved in phospholipid metabolism and immune responses, monoglyceride lipase (Mgll; ~1.8-fold) involved in triglyceride hydrolysis, and fatty acid uptake and transport related genes for very low density lipoprotein receptor (Vldlr; ~2.1-fold), Cd36 (~3.9-fold), and fatty acid binding proteins (Fabps) 4, 5 and 12 (~1.5-, ~2.2-, and ~22-fold, respectively). Several of these genes have 19 bp DRE sequences with an MSS>0.8 in the 10 kb TSS upstream region (Agpat3, Cd36, Lpcat1, Pla2g15, Alox12, Fabp4, Fabp5; Supplementary Table S3). Furthermore, pDREs>0.8473 are found in the mouse Fabp12 (MSS of 0.873 and 0.894) and Mgll (MSS of 0.857) genomic regions along with AhR enrichment which is consistent with TCDD-mediated induction. However, mouse Vldlr shows AhR enrichment in the absence of any high-scoring DRE sequences, suggesting activation by non-canonical mechanisms (Beischlag et al., 2008; Denison et al., 2011; Dere et al., 2011c; Huang and Elferink, 2012).
Table 1.
Over-represented functions associated with species-specific differential gene expression.
| Category | Enrichment score |
|---|---|
| Rat | |
| Antigen processing and presentation | 6.15 |
| Nucleotide binding | 2.42 |
| Acetyltransferase activity | 1.84 |
| Mouse | |
| Inflammatory response | 6.79 |
| Chemotaxis | 5.52 |
| Immune response | 5.48 |
| Response to bacteria | 4.96 |
| Immune cell activation and development | 3.71 |
| Antigen processing and presentation | 3.43 |
| Cytokine activity | 3.34 |
| Oxidation/reduction | 3.32 |
| Cytokine production | 3.28 |
| Enzyme inhibitor activity | 3.02 |
| Steroid metabolism | 2.79 |
| Antigen receptor-mediated signaling | 2.05 |
| Immune cell proliferation | 2.03 |
| Xenobiotic metabolism | 2.02 |
| Bone development | 1.88 |
| Carbohydrate binding | 182 |
| Phagocytosis | 1.66 |
| Fatty acid metabolism | 1.57 |
| Lipid catabolism | 1.5 |
| Angiogenesis | 1.46 |
| Alcohol biosynthesis | 1.45 |
| Glycerolipid metabolism | 1.35 |
Transcription factor analysis
Gene expression datasets were used to investigate species-specific transcription factor activation or inhibition. A total of 22 rat and 40 mouse transcriptional regulators (TRs) were identified (Table 2), 9 of which were predicted to be active in both species. The glucocorticoid receptor (GR), interferon regulatory factors (IRF) 1, 3, and 7, and nuclear factor (erythroid-derived 2)-like 2 (NFE2L2, also known as NRF2) were predicted to show divergent activity. GR and IRF are involved in stress and immune responses (Nunez et al., 2005) and were predicted to be activated in rat while inhibited in mouse. In contrast, NFE2L2, an important oxidative stress signaling mediator shown to be activated by TCDD (Boutros et al., 2009), was predicted to be inhibited in rat and activated in mouse. The predicted mouse-specific induction of NFE2L2 activity is consistent with mouse-specific gene expression induction of Nqo1, Ugts, and Gsts, members of the “TCDD-inducible AhR-Nrf2 gene battery” (Yeager et al., 2009). Although the predicted down-regulation of thyroid hormone receptor β (THRB) activity in both species is consistent with hypothyroxemia observed in rats exposed to TCDD, hypothyroxemia is not reported in mice (Craft et al., 2002). Other mouse-specific TR predictions include the activation of the AhR, repression of the carbohydrate response element binding protein (ChREBP), and repression of the peroxisome-proliferator activated receptor (PPAR) δ and γ activities. Although TR activation or inhibition may be post-translational, ChREBP mRNA exhibited a corresponding ~1.6-fold inhibition of expression.
Table 2.
Transcription factor analysis of rat and mouse gene sets.
| Transcription regulator | Rat |
Mouse |
||||
|---|---|---|---|---|---|---|
| 1d | 3d | 5d | 1d | 3d | 7d | |
| AhR | – | – | – | ▲ – | ▲ – | – |
| C/EBPa | ▼ – | – | – | –1.56 | – | – |
| CAR | – | – | – | 1.71 | ▲ 1.7 | – |
| ChREBP | – | – | – | ▼ – 1.59 | ▼ – | ▼ – |
| CLOCK | ▲ – | – | ▲ – | – | – | – |
| Creb | – | – | – | – | – | ▼ – |
| FOXG1 | – | – | – | ▼ – | – | – |
| GATA2 | – | – | – | ▼ – | – | – |
| GR | NA | ▲ NA | NA | – | – | ▼ – |
| HNF1A | – | – | – | ▼ – | ▼ – | ▼ – |
| HSF1 | – | – | – | – | – | ▼ – |
| IRF1 | – | ▲ – | – | – | – | ▲ – |
| IRF3 | – | ▲ – | – | ▼ – | ▼ – | – |
| IRF7 | – | ▲ 2.56 | – | ▼ –1.69 | – | – |
| KEAP1 | – | – | – | – | ▼ – | – |
| LXRA | – | – | – | – | – | ▼ – |
| LXRB | NA | ▼ NA | NA | – | – | – |
| MLX | – | – | – | ▼ – | ▼ – | – |
| MR | – | – | – | – | – | ▲ – |
| MYC | – | – | – | 2.04 | 1.55 | ▼ 2.2 |
| NFE2L2 | – | ▼ – | – | ▲ 3.51 | ▲ 2.38 | 2.45 |
| NFkB | – | – | – | – | – | – |
| NFYA | – | ▼ – | – | – | – | – |
| NOTCH1 | – | – | – | 3.11 | ▲ 2.86 | ▲ 2.97 |
| NRIP1 | – | –1.72 | – | – | – | ▲ 1.51 |
| PPARγ | –1.61 | – | – | – | – | ▼ – |
| PPAR8 | – | –2.04 | – | – | – | ▼ – |
| RB1 | – | ▲ – | – | – | – | – |
| RELA | – | – | – | – | – | ▲ – |
| RXRA | – | – | ▼ – | – | – | – |
| SKIL | NA | ▼ NA | NA | – | – | ▼ – |
| SMAD2 | – | – | – | ▲ – | – | – |
| SMAD3 | – | – | – | ▲ – | ▲ – | – |
| SP3 | – | – | – | – | ▲ – | ▲ – |
| SPDEF | – | ▲ – | – | – | – | – |
| SREBF1 | – | –1.96 | – | –1.67 | ▼ – 1.64 | ▼ – 1.56 |
| SREBF2 | – | – | – | – | ▼ – | ▼ – |
| STAT1 | NA | ▲ NA | NA | – | – | ▲ 1.53 |
| STAT2 | – | ▲ – | – | – | – | – |
| STAT4 | – | ▲ – | – | – | ▲ – | – |
| TBX2 | – | ▼ – | – | – | – | – |
| TBX21 | NA | NA | NA | – | – | ▲ 1.63 |
| TCF3 | – | ▲ – | – | – | – | – |
| THRB | – | ▼ – | ▼ – | – | – | ▼ – |
| TOB1 | – | – | – | – | – | ▼ – |
| TP53 | – | ▲ – | – | |||
| TRIM24 | NA | NA | NA | ▲ – | – | – |
| XBP1 | – | – | – | – | ▼ – | ▼ – |
Note: arrows indicate predicted activation (▲)/inhibition (▼) with a |z-score| ≥2 by transcription factor analysis in IPA. Numbers represent gene expression values (|fold change| ≥ 1.5, P1(t) ≥0.99). Lines (–) indicate no change in mRNA expression of the TR meeting filtering criteria and NA indicates una ▲ lable gene expression data.
Discussion
Species-specific toxicogenomic and histological responses elicited by TCDD have been reported in rats and mice (Boutros et al., 2008; Boverhof et al., 2006; Carlson et al., 2009; Dere et al., 2011b; Forgacs et al., 2012, submitted for publication; Poland and Knutson, 1982). More specifically, comparative in vivo rat and mouse toxicogenomic studies suggest that <20% of represented orthologs exhibit differential expression in both species using arrays with limited genomic coverage (Boutros et al., 2008; Boverhof et al., 2006). We have extended this comparative analysis to 11,708 orthologous genes using rat Affymetrix (DrugMatrix) and mouse Agilent (Dere et al., 2011c) datasets identifying only 70 commonly regulated orthologs representing <15% of TCDD-responsive genes in either species.
Although the datasets used in this study were generated using different study designs, treatment regimens, and microarray platforms, valid inter-platform comparisons are possible when raw signal intensities are re-analyzed using the same normalization and statistical methods (Guo et al., 2006; Shi et al., 2006). More importantly, re-annotation of the rat DrugMatrix dataset was required for accurate comparisons as updated annotation changed the gene association for ~29% of the probe sets. Nevertheless, using a similar fold-change threshold as in previous species comparison (Dere et al., 2011b) the relative number of species-specific and conserved differentially expressed orthologs was consistent with previous comparative studies (Boutros et al., 2008; Boverhof et al., 2006; Dere et al., 2011b). Moreover, b20% of TCDD-responsive orthologs are conserved between species using the more relaxed filtering criteria (data not shown), similar to previous reports (Boutros et al., 2008; Boverhof et al., 2006).
The large number of species-specific responses is also consistent with the divergent DRE distributions in the regulatory regions of differentially expressed orthologs (Dere et al., 2011a,c; Sun et al., 2004). Despite distinct TCDD-responsive orthologs, both rat and mouse were significantly enriched for antigen processing and presentation (most of which showed up-regulation) implicating induced immune responses in both species. Histological examination also reveals immune cell infiltration in mouse liver (Boverhof et al., 2006) and inflammation in rat liver (National Toxicology, 2006). Conversely, rat-specific effects on nucleotide binding and acetyltransferase activity suggest dynamic transcriptional regulation. Rat-specific TCDD-mediated induction of phospholipid metabolism-related Agpat3 and Lpcat1 genes, and the repression of Gpam and Pla2g15 may also be involved in the dysregulation of choline metabolism in rats (Forgacs et al., 2012).
Acute TCDD exposure elicits mouse-specific development of hepatic steatosis (Boverhof et al., 2006). Mouse-specific induction of Fabp4, 5, and 12, Vldlr, and Cd36 is indicative of increased fatty acid uptake and transport, and consistent with dietary fat as the principle source of lipids in TCDD-elicited steatosis (Angrish et al., 2012; Atshaves et al., 2010; Goudriaan et al., 2001). The absence of pDREs in the 10 kb TSS upstream region for Vldlr suggests the involvement of non-canonical mechanisms in hepatic steatosis induced by TCDD. The induction of lipases, including Lpl, further supports free fatty acid uptake from hydrolyzed lipoproteins while the down-regulation of Ceacam1 (Boverhof et al., 2006; DeAngelis et al., 2008), and the induction of Mgll induction are consistent with increased hydrolysis of triglyceride stores and hepatic fatty acid accumulation in the mouse. In contrast, the lack of rat hepatic lipid accumulation may be due to AhR-mediated Ceacam1 induction consistent with the presence of pDREs in the regulatory region. While the down-regulation of Pnpla3 in both species should facilitate hepatic triglyceride accumulation (He et al., 2010), it is contrary to the elicited hepatic steatosis reported in the mouse but not in the rat following a single bolus oral dose of 10 μg/kg TCDD (Boverhof et al., 2006).
Mouse-specific predictions of PPARδ, PPARγ, and ChREBP inhibition also indicate TCDD-mediated metabolic disruption. Interestingly, TCDD-elicited differential gene expression in rat and human primary hepatocytes identified human-specific enrichment of PPAR and ChREBP signaling pathways (Black et al., 2012). The absence of predicted PPAR and ChREBP activity in the rat suggests that human responses associated with energy homeostasis may be more similar to mice. However, ChREBP inhibition alleviated hepatic steatosis in ob/ob mice (Postic et al., 2007), contrary to lipid accumulation in TCDD-treated mouse liver, highlighting the importance of considering whole-genome data and ‘toxicity pathway’ elucidation, as opposed to individual gene expression responses.
In summary, despite increasing the ortholog coverage by using the rat Affymetrix dataset available within DrugMatrix, only a small number of orthologs are regulated by TCDD in the mouse and rat, despite using similar normalization and statistical approaches as well as the most current annotation. Several differentially expressed orthologs associated with fatty acid, glycerolipid, and carbohydrate metabolism emphasize species-specific metabolic responses consistent with TCDD-mediated hepatic steatosis in the mouse (Boverhof et al., 2006) and perturbations of choline metabolism in rat (Forgacs et al., 2012). These results provide further evidence that TCDD elicits species-specific responses by regulating distinct gene networks. However, it remains unclear which rodent is a better model for humans although comparative studies in primary hepatocytes suggest that mice may be a better model for selected responses such as hepatic fat accumulation (Forgacs et al., submitted for publication). Further assessment of species-specific regulated gene networks is warranted to identify other conserved responses/pathways relevant to human health.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.taap.2012.11.028.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences Superfund Basic Research Program (NIEHS SBRP P42ES04911). The authors would like to thank Agnes Forgacs and Dr. Anna Kopec for the critical review of this manuscript as well as Dr. Scott Auerbach for providing raw data and technical support for the DrugMatrix database.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- Angrish MM, Mets BD, Jones AD, Zacharewski TR. Dietary fat is a lipid source in 2,3,7,8-tetrachlorodibenzo-rho-dioxin (TCDD)-elicited hepatic steatosis in C57BL/6 mice. Toxicol. Sci. 2012;128:377–386. doi: 10.1093/toxsci/kfs155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atshaves BP, Martin GG, Hostetler HA, McIntosh AL, Kier AB, Schroeder F. Liver fatty acid-binding protein and obesity. J. Nutr. Biochem. 2010;21:1015–1032. doi: 10.1016/j.jnutbio.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank PA, Yao EF, Phelps CL, Harper PA, Denison MS. Species-specific binding of transformed Ah receptor to a dioxin responsive transcriptional enhancer. Eur. J. Pharmacol. 1992;228:85–94. doi: 10.1016/0926-6917(92)90016-6. [DOI] [PubMed] [Google Scholar]
- Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydro-carbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black MB, Budinsky RA, Dombkowski A, Cukovic D, LeCluyse EL, Ferguson SS, Thomas RS, Rowlands JC. Cross-species comparisons of transcriptomic alterations in human and rat primary hepatocytes exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 2012;127:199–215. doi: 10.1093/toxsci/kfs069. [DOI] [PubMed] [Google Scholar]
- Boutros PC, Bielefeld KA, Pohjanvirta R, Harper PA. Transcriptomic responses to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in liver: comparison of rat and mouse. BMC Genomics. 2008;9:419. doi: 10.1186/1471-2164-9-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros PC, Yan R, Moffat ID, Pohjanvirta R, Okey AB. Dioxin-dependent and dioxin-independent gene batteries: comparison of liver and kidney in AHR-null mice. Toxicol. Sci. 2009;112:245–256. doi: 10.1093/toxsci/kfp191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Chittim B, Harkema JR, Jump DB, Zacharewski TR. Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-mediated hepatotoxicity. Toxicol. Sci. 2005;85:1048–1063. doi: 10.1093/toxsci/kfi162. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Sharratt B, Chittim B, Harkema JR, Mendrick DL, Zacharewski TR. Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 mice. Toxicol. Sci. 2006;94:398–416. doi: 10.1093/toxsci/kfl100. [DOI] [PubMed] [Google Scholar]
- Budinsky RA, LeCluyse EL, Ferguson SS, Rowlands JC, Simon T. Human and rat primary hepatocyte CYP1A1 and 1A2 induction with 2,3,7,8-tetrachlorodibenzo-pdioxin, 2,3,7,8-tetrachlorodibenzofuran, and 2,3,4,7,8-pentachlorodibenzofuran. Toxicol. Sci. 2010;118:224–235. doi: 10.1093/toxsci/kfq238. [DOI] [PubMed] [Google Scholar]
- Burgoon LD, Boutros PC, Dere E, Zacharewski TR. dbZach: a MIAME-compliant toxicogenomic supportive relational database. Toxicol. Sci. 2006;90:558–568. doi: 10.1093/toxsci/kfj097. [DOI] [PubMed] [Google Scholar]
- Carlson EA, McCulloch C, Koganti A, Goodwin SB, Sutter TR, Silkworth JB. Divergent transcriptomic responses to aryl hydrocarbon receptor agonists between rat and human primary hepatocytes. Toxicol. Sci. 2009;112:257–272. doi: 10.1093/toxsci/kfp200. [DOI] [PubMed] [Google Scholar]
- Craft ES, DeVito MJ, Crofton KM. Comparative responsiveness of hypothyroxinemia and hepatic enzyme induction in Long–Evans rats versus C57BL/6J mice exposed to TCDD-like and phenobarbital-like polychlorinated biphenyl congeners. Toxicol. Sci. 2002;68:372–380. doi: 10.1093/toxsci/68.2.372. [DOI] [PubMed] [Google Scholar]
- DeAngelis AM, Heinrich G, Dai T, Bowman TA, Patel PR, Lee SJ, Hong EG, Jung DY, Assmann A, Kulkarni RN, Kim JK, Najjar SM. Carcinoembryonic antigen-related cell adhesion molecule 1 — a link between insulin and lipid metabolism. Diabetes. 2008;57:2296–2303. doi: 10.2337/db08-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison MS, Heath-Pagliuso S. The Ah receptor: a regulator of the biochemical and toxicological actions of structurally diverse chemicals. Bull. Environ. Contam. Toxicol. 1998;61:557–568. doi: 10.1007/pl00002973. [DOI] [PubMed] [Google Scholar]
- Denison MS, Vella LM, Okey AB. Structure and function of the Ah receptor for 2,3,7,8-tetrachlorodibenzo-p-dioxin. Species difference in molecular properties of the receptors from mouse and rat hepatic cytosols. J. Biol. Chem. 1986;261:3987–3995. [PubMed] [Google Scholar]
- Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci. 2011;124:1–22. doi: 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dere E, Forgacs AL, Zacharewski TR, Burgoon LD. Genome-wide computational analysis of dioxin response element location and distribution in the human, mouse, and rat genomes. Chem. Res. Toxicol. 2011a;24:494–504. doi: 10.1021/tx100328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E, Lee AW, Burgoon LD, Zacharewski TR. Differences in TCDD-elicited gene expression profiles in human HepG2, mouse Hepa1c1c7 and rat H4IIE hepatoma cells. BMC Genomics. 2011b;12:193. doi: 10.1186/1471-2164-12-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E, Lo R, Celius T, Matthews J, Zacharewski TR. Integration of genome-wide computation DRE search, AhR ChIP-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC Genomics. 2011c;12:365. doi: 10.1186/1471-2164-12-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel JE, Gennings C, Chinchilli VM, Burgoon LD, Zacharewski TR. Empirical Bayes gene screening tool for time-course or dose–response microarray data. J. Biopharm. Stat. 2004;14:647–670. doi: 10.1081/BIP-200025656. [DOI] [PubMed] [Google Scholar]
- Eckel JE, Gennings C, Therneau TM, Burgoon LD, Boverhof DR, Zacharewski TR. Normalization of two-channel microarray experiments: a semiparametric approach. Bioinformatics. 2005;21:1078–1083. doi: 10.1093/bioinformatics/bti105. [DOI] [PubMed] [Google Scholar]
- Fletcher N, Wahlstrom D, Lundberg R, Nilsson CB, Nilsson KC, Stockling K, Hellmold H, Hakansson H. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) alters the mRNA expression of critical genes associated with cholesterol metabolism, bile acid biosynthesis, and bile transport in rat liver: a microarray study. Toxicol. Appl. Pharmacol. 2005;207:1–24. doi: 10.1016/j.taap.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Forgacs AL, Kent MN, Makley MK, Mets B, DelRaso N, Jahns GL, Burgoon LD, Zacharewski TR, Reo NV. Comparative metabolomic and genomic analyses of TCDD-elicited metabolic disruption in mouse and rat liver. Toxicol. Sci. 2012;125:41–55. doi: 10.1093/toxsci/kfr262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgacs AL, Dere E, Angrish MM, Zacharewski T. Comparative analysis of TCDD-elicited gene expression in human, mouse and rat primary hepatocytes. doi: 10.1093/toxsci/kft028. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, Diekhans M, Dreszer TR, Giardine BM, Harte RA, Hillman-Jackson J, Hsu F, Kirkup V, Kuhn RM, Learned K, Li CH, Meyer LR, Pohl A, Raney BJ, Rosenbloom KR, Smith KE, Haussler D, Kent WJ. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganter B, Tugendreich S, Pearson CI, Ayanoglu E, Baumhueter S, Bostian KA, Brady L, Browne LJ, Calvin JT, Day GJ, Breckenridge N, Dunlea S, Eynon BP, Furness LM, Ferng J, Fielden MR, Fujimoto SY, Gong L, Hu C, Idury R, Judo MS, Kolaja KL, Lee MD, McSorley C, Minor JM, Nair RV, Natsoulis G, Nguyen P, Nicholson SM, Pham H, Roter AH, Sun D, Tan S, Thode S, Tolley AM, Vladimirova A, Yang J, Zhou Z, Jarnagin K. Development of a large-scale chemogenomics database to improve drug candidate selection and to understand mechanisms of chemical toxicity and action. J. Biotechnol. 2005;119:219–244. doi: 10.1016/j.jbiotec.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Fernandez-Salguero P. The aryl hydrocarbon receptor: studies using the AHR-null mice. Drug Metab. Dispos. 1998;26:1194–1198. [PubMed] [Google Scholar]
- Gonzalez FJ, Shah YM. PPARalpha: mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology. 2008;246:2–8. doi: 10.1016/j.tox.2007.09.030. [DOI] [PubMed] [Google Scholar]
- Goudriaan JR, Tacken PJ, Dahlmans VEH, Gijbels MJJ, van Dijk KW, Havekes LM, Jong MC. Protection from obesity in mice lacking the VLDL receptor. Arterioscler. Thromb. Vasc. Biol. 2001;21:1488–1493. doi: 10.1161/hq0901.095147. [DOI] [PubMed] [Google Scholar]
- Guo L, Lobenhofer EK, Wang C, Shippy R, Harris SC, Zhang L, Mei N, Chen T, Herman D, Goodsaid FM, Hurban P, Phillips KL, Xu J, Deng X, Sun YA, Tong W, Dragan YP, Shi L. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat. Biotechnol. 2006;24:1162–1169. doi: 10.1038/nbt1238. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Karchner SI, Shapiro MA, Perera SA. Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13743–13748. doi: 10.1073/pnas.94.25.13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- He S, McPhaul C, Li JZ, Garuti R, Kinch L, Grishin NV, Cohen JC, Hobbs HH. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J. Biol. Chem. 2010;285:6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Elferink CJ. A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol. Pharmacol. 2012;81:338–347. doi: 10.1124/mol.111.075952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPA [February 20, 2012];Ingenuity Transcription Factor Analysis in IPA [White Paper] 2011 https://ingenuity-c.na3.content.force.com/servlet/fileField?id=0BE50000000Gmim.
- Kim S, Dere E, Burgoon LD, Chang CC, Zacharewski TR. Comparative analysis of AhR-mediated TCDD-elicited gene expression in human liver adult stem cells. Toxicol. Sci. 2009;112:229–244. doi: 10.1093/toxsci/kfp189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec AK, Boverhof DR, Burgoon LD, Ibrahim-Aibo D, Harkema JR, Tashiro C, Chittim B, Zacharewski TR. Comparative toxicogenomic examination of the hepatic effects of PCB126 and TCDD in immature, ovariectomized C57BL/6 mice. Toxicol. Sci. 2008;102:61–75. doi: 10.1093/toxsci/kfm289. [DOI] [PubMed] [Google Scholar]
- Kopec AK, D'Souza ML, Mets BD, Burgoon LD, Reese SE, Archer KJ, Potter D, Tashiro C, Sharratt B, Harkema JR, Zacharewski TR. Non-additive hepatic gene expression elicited by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and 2,2′,4,4′,5,5′-hexachlorobiphenyl (PCB153) co-treatment in C57BL/6 mice. Toxicol. Appl. Pharmacol. 2011;256:154–167. doi: 10.1016/j.taap.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. The CEACAM1 expression is decreased in the liver of severely obese patients with or without diabetes. Diagn. Pathol. 2011;6:40. doi: 10.1186/1746-1596-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane×receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol. Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- Najjar SM. Regulation of insulin action by CEACAM1. Trends Endocrinol. Metab. 2002;13:240–245. doi: 10.1016/s1043-2760(02)00608-2. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program NTP technical report on the toxicology and carcinogenesis studies of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (CAS No. 1746-01-6) in female Harlan Sprague–Dawley rats (gavage studies). National Toxicology Program Technical Report Series. 2006:4–232. [PubMed] [Google Scholar]
- NIEHS/NTP [October 4, 2012];DrugMatrix Calculations [White Paper] 2011 https://ntp.niehs.nih.gov/drugmatrix/projects/DrugMatrix/support/White_Paper/DrugMatrix_Calculations.pdf.
- Nunez BS, Geng C-D, Pedersen KB, Millro-Macklin CD, Vedeckis WV. Interaction between the interferon signaling pathway and the human glucocorticoid receptor gene 1A promoter. Endocrinology. 2005;146:1449–1457. doi: 10.1210/en.2004-0672. [DOI] [PubMed] [Google Scholar]
- Okey AB, Riddick DS, Harper PA. Molecular biology of the aromatic hydro-carbon (dioxin) receptor. Trends Pharmacol. Sci. 1994;15:226–232. doi: 10.1016/0165-6147(94)90316-6. [DOI] [PubMed] [Google Scholar]
- Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W, Dorato M, Van Deun K, Smith P, Berger B, Heller A. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul. Toxicol. Pharmacol. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- Poland A, Knutson JC. 2,3,7,8-Tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Postic C, Dentin R, Denechaud PD, Girard J. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu. Rev. Nutr. 2007;27:179–192. doi: 10.1146/annurev.nutr.27.061406.093618. [DOI] [PubMed] [Google Scholar]
- Shi L, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth JB, Koganti A, Illouz K, Possolo A, Zhao M, Hamilton SB. Comparison of TCDD and PCB CYP1A induction sensitivities in fresh hepatocytes from human donors, Sprague–Dawley rats, and rhesus monkeys and HepG2 cells. Toxicol. Sci. 2005;87:508–519. doi: 10.1093/toxsci/kfi261. [DOI] [PubMed] [Google Scholar]
- Smith LL. Key challenges for toxicologists in the 21st century. Trends Pharmacol. Sci. 2001;22:281–285. doi: 10.1016/s0165-6147(00)01714-4. [DOI] [PubMed] [Google Scholar]
- Sun YV, Boverhof DR, Burgoon LD, Fielden MR, Zacharewski TR. Comparative analysis of dioxin response elements in human, mouse and rat genomic sequences. Nucleic Acids Res. 2004;32:4512–4523. doi: 10.1093/nar/gkh782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. (RESEARCH0034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager RL, Reisman SA, Aleksunes LM, Klaassen CD. Introducing the “TCDD-inducible AhR–Nrf2 gene battery”. Toxicol. Sci. 2009;111:238–246. doi: 10.1093/toxsci/kfp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.