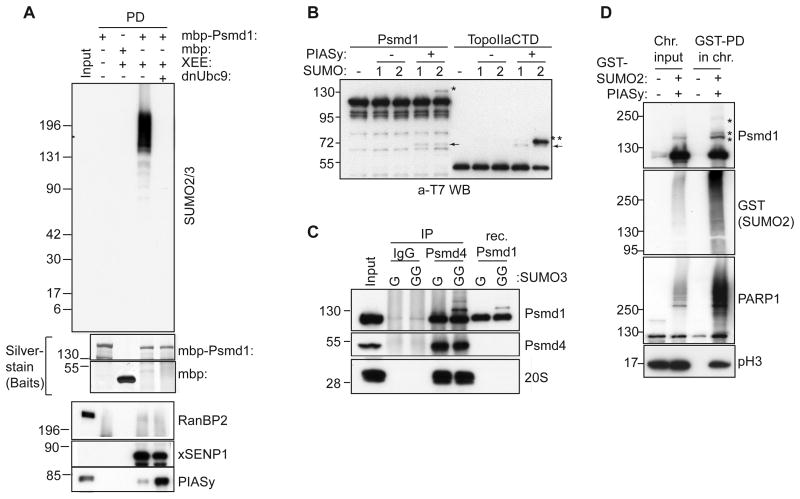
Figure 2. Psmd1 is modified by SUMO2/3 in XEE and in vitro.
(A) MBP or MBP-Psmd1 were incubated under the indicated conditions, and pulled down using Amylose resin. Where indicated, dominant negative E2 (dnUbc9) was included to inhibit SUMOylation. The samples were analyzed by Western blotting with antibodies against the indicated proteins. Silver stain shows bait proteins. Input: 5%
(B) T7-tagged Psmd1 or T7-tagged TopoIIαCTD were subjected to in vitro SUMOylation with or without PIASy, and analyzed by Western blotting with anti-T7 antibodies. 1 and 2 indicate SUMO1 and SUMO2, respectively. Asterisks and arrows indicate SUMO-conjugated species and PIASy, respectively.
(C) Immunoprecipitations (IP) from XEE using IgG and anti-Psmd4 antibodies were subjected to in vitro reactions as in (B), except that SUMO3 replaced the other paralogs. GG indicates that the mature form of SUMO3 was used, while G indicates use of a truncated, non-conjugatable form. Recombinant (Rec.) Psmd1 was concurrently subjected to in vitro SUMOylation. The samples were analyzed by Western blotting, as indicated. Antibodies against subunit C2 were used to detect 20S proteasome. Input: 5%
(D) Sperm chromatin was incubated for 60 min in XEE in the absence or presence of GST-SUMO2 and PIASy. The isolated chromosome fractions (Chr.) were processed and pulled down for GST-SUMO2 (see experimental procedures), followed by Western blotting with the indicated antibodies. Asterisks indicate SUMO-modified forms of Psmd1. pH3 indicates phosphohistone H3. Input: 6%