Fig. 2.
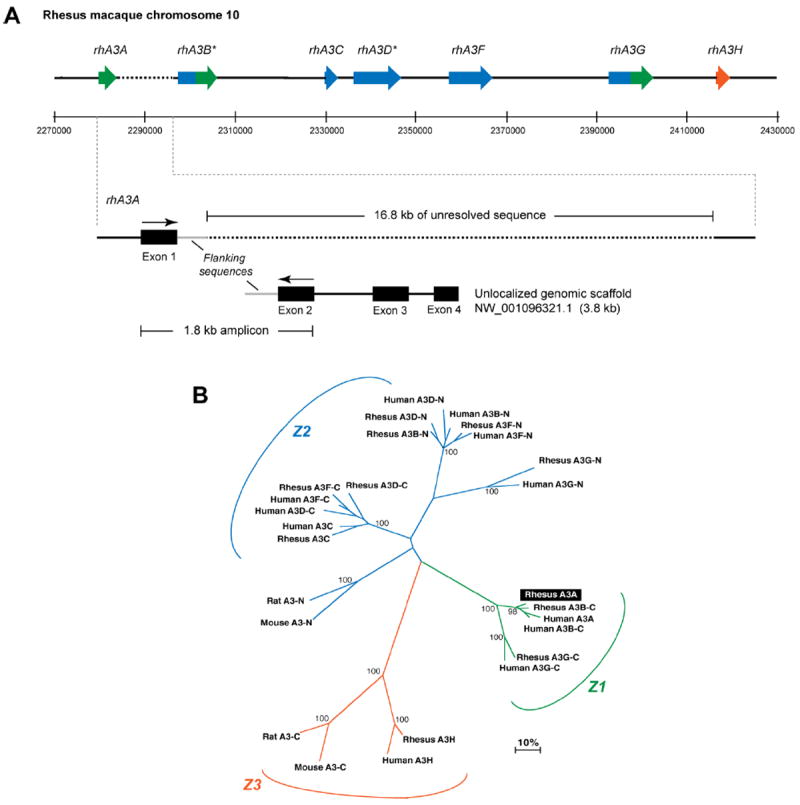
Sequence characterization of the rhesus macaque A3 locus. Panel A. Genome organization of rhesus macaque A3 genes. (Top) Published cDNA sequences for rhA3C, rhA3F, rhA3G and rhA3H were subjected to BLAST analysis against the rhesus macaque genome. Matching exons were plotted against assigned genome coordinates, shown here in 20 kb intervals. *RhA3B and rhA3DE were predicted by subjecting the genome sequences upstream of rhA3C or flanked by rhA3C and rhA3F to the geneid prediction program (http://genome.crg.es/geneid.html), respectively. RhA3A amplified from activated PBMCs were subjected to BLAST analysis. RhA3A exon 1 mapped in the genome directly upstream of A3B, but this was followed by 16.8 kb of unresolved sequence (dotted lines). (Bottom) A3A exons 2 to 4 matched sequences from an unlocalized chromosome 10 genomic scaffold sequence. Primers based on A3A exons 1 and 2 amplified a 1.8 kb product that matched flanking sequences after exon 1 in the genome and before exon 2 in the unlocalized genomic scaffold (gray lines). This provides strong evidence for the current placement of rhA3A in the rhesus macaque genome. Panel B. Phylogeny of rhesus macaque and human A3 proteins. Amino acid sequences from rhesus macaque and human A3 genes were aligned, with the double-CDA members split into an N- and C-terminal half. For example, A3B-C corresponds to the C-terminal half of A3B. RhA3A clustered with the C-terminal half of A3B and A3G, forming the Z1 clade. Each rhesus macaque A3 gene clustered significantly with the corresponding human A3 gene, suggesting that rhesus macaque and human A3 proteins are orthologous.
