Significance
The body stores excess blood glucose as glycogen, a sugary substance that contains up to 55,000 glucose molecules joined together as a chain, mostly in liver and muscle cells. Conversion of glucose to glycogen and glycogen to glucose in these cells plays an important role in regulating blood glucose levels. Glycogen ensures that we don’t run out of fuel during prolonged exercise. To make glycogen from blood sugar, cells need two enzymes: glycogenin and glycogen synthase. Glycogenin kick starts the process by first linking to itself a string of glucose residues and then recruiting glycogen synthase to elaborate this “seed” glycogen particle. Here, we describe the molecular details of how these two enzymes come together and begin to make glycogen.
Keywords: glucose metabolism, energy metabolism, glycogenesis, starch
Abstract
Glycogen is a primary form of energy storage in eukaryotes that is essential for glucose homeostasis. The glycogen polymer is synthesized from glucose through the cooperative action of glycogen synthase (GS), glycogenin (GN), and glycogen branching enzyme and forms particles that range in size from 10 to 290 nm. GS is regulated by allosteric activation upon glucose-6-phosphate binding and inactivation by phosphorylation on its N- and C-terminal regulatory tails. GS alone is incapable of starting synthesis of a glycogen particle de novo, but instead it extends preexisting chains initiated by glycogenin. The molecular determinants by which GS recognizes self-glucosylated GN, the first step in glycogenesis, are unknown. We describe the crystal structure of Caenorhabditis elegans GS in complex with a minimal GS targeting sequence in GN and show that a 34-residue region of GN binds to a conserved surface on GS that is distinct from previously characterized allosteric and binding surfaces on the enzyme. The interaction identified in the GS-GN costructure is required for GS–GN interaction and for glycogen synthesis in a cell-free system and in intact cells. The interaction of full-length GS-GN proteins is enhanced by an avidity effect imparted by a dimeric state of GN and a tetrameric state of GS. Finally, the structure of the N- and C-terminal regulatory tails of GS provide a basis for understanding phosphoregulation of glycogen synthesis. These results uncover a central molecular mechanism that governs glycogen metabolism.
Glycogen forms the major rapidly accessible energy reserve in eukaryotes and, as such, is essential for cellular and whole-body energy supply and glucose homeostasis. In mammals, glucose is stored as glycogen mainly in muscle and liver cells (and to a lesser extent in astrocytes, adipocytes, and kidney and pancreatic cells) when blood glucose levels are high, and then released for utilization within the cell, or systemically when glucose and energy levels are low. The dysregulation of glycogen metabolism contributes to glycogen storage diseases (1), cardiac myopathies (2), neurodegeneration (3), insulin resistance (4), and cancer (5). Notably, the up-regulation of glycogen synthesis provides an alternate source of energy under hypoxic conditions and contributes to cancer cell survival in preangiogenic states (5).
Glycogen is a branched polymer of glucose formed primarily through α1,4 glycosidic linkages, with periodic intersecting α1,6 linkages serving as branch points. In eukaryotes, glycogen is synthesized through the cooperative action of three enzymes, namely glycogen synthase (GS), glycogenin (GN), and glycogen branching enzyme (GBE) (1), which use UDP-glucose (UDP-G) as a glucose donor. A GN dimer initiates the glycogen polymer by autoglucosylation of a conserved tyrosine residue (Tyr195 in human GN1, Tyr230 in yeast GN1, and Tyr194 in Caenorhabditis elegans GN), leading to an α1,4-linked chain of 8–12 glucose units (6). This oligosaccharide remains attached to glycogenin and forms a primer that is converted into a full-size glycogen particle by the combined actions of GS and GBE (1, 7). Once fully elaborated, a glycogen particle can comprise up to ∼55,000 glucose residues with a size distribution in muscle tissue of 10–44 nm in diameter, termed β particles (8, 9). In liver, glycogen particles of 110–290 nm in diameter, termed α particles, are formed by the assembly of several β particles, possibly through covalent linkage (9, 10). Glycogen particle size varies greatly between tissues and species (11), but the basis and significance of these size differences are poorly understood.
Glycogenin is a member of the GT8 family of glycosyltransferases with a GT-A architecture containing an N-terminal catalytic domain with a single Rossmann fold that operates as an obligate dimer (12–14). The core catalytic domain is followed by a C-terminal extension of variable length and undefined structure. The last 35 amino acids of this tail in human and yeast contain a conserved motif that is sufficient for binding to GS in cell lysates (15). The region that separates the core catalytic domain of GN and the GS binding motif is highly variable both in sequence and in length (Fig. 1A). Most eukaryotes possess two versions of GN that differ in the length of this linker, which ranges from 50 to 257 residues in yeast, 7–142 residues in worm and 34–170 residues in human. In addition, the linker region is a site of alternative splicing in humans that imparts further length variation (16). The functional significance of the variability in linker length has yet to be explored.
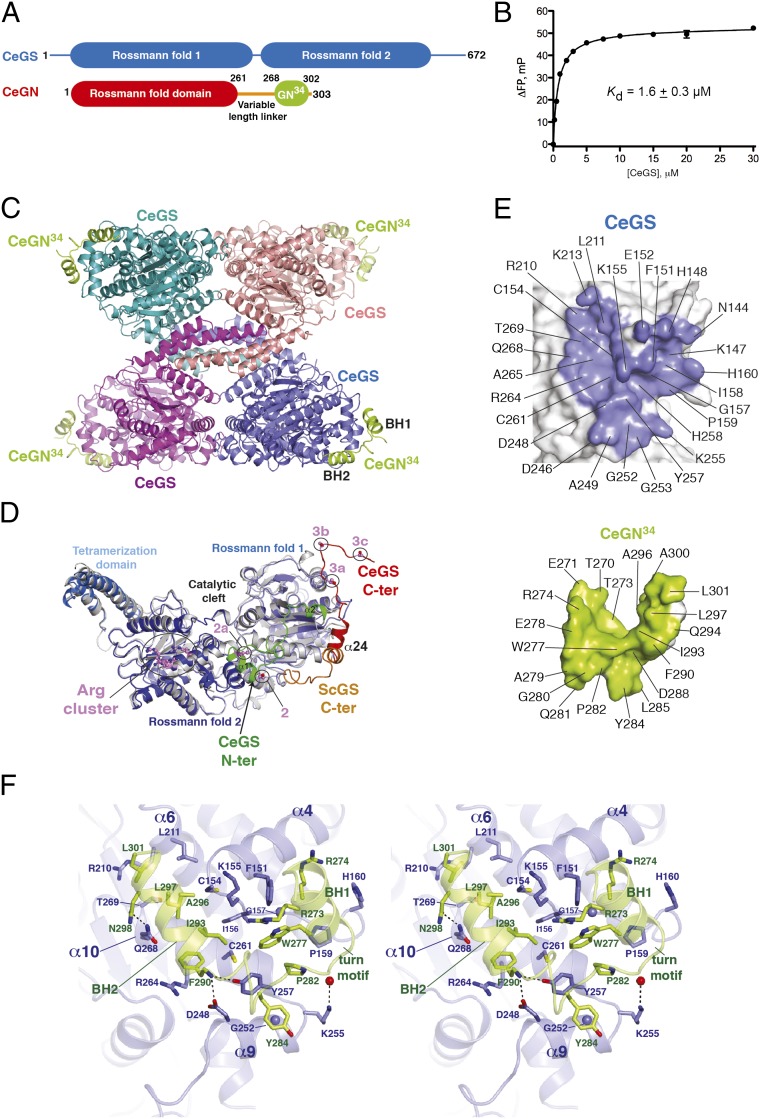
Fig. 1.
Structure of CeGS in complex with CeGN34. (A) Domain architecture of CeGS (GT-B fold) and CeGN (GT-A fold). (B) Interaction of CeGN34 to CeGS determined by fluorescence polarization. Kd value ± SEM is the average of three independent experiments carried out in duplicate. (C) Structure of the CeGS tetramer bound to the CeGN34 peptide. (D) Comparison of CeGS protomers (blue) and the budding yeast ScGS (PDB ID code 3NAZ, gray). The N-terminal extension of CeGS (green) and C-terminal extensions of CeGS (red) and ScGS (orange) are highlighted. Phosphoregulatory sites 2 (S12), 2a (T19), 3a (S654), 3b (S658), and 3c (S662) and the allosteric regulatory site (Arg cluster) on the CeGS protomer are shown as ball and stick models with violet-colored carbon atoms. (E) Peel away surface representations of the CeGS–CeGN complex with contact residues (CeGS, Upper; CeGN, Lower). (F) Detailed stereoview of the CeGS–CeGN34 complex. CeGS is shown in blue and CeGN in green. Side chain residues that make direct contacts are shown as sticks with colored heteroatoms (oxygen, red; nitrogen, dark blue; sulfur, yellow). Glycine residues are depicted as blue spheres, and carbonyl oxygens are shown as red spheres. Hydrogen bonds are depicted as dotted lines.
GS synthesizes α1,4-linked glucose polymers. The mammalian and yeast GS enzymes belong to the GT3 family of glycosyltransferases and are regulated by covalent phosphorylation and allosteric ligand interactions. In contrast, the bacterial and plant GS enzymes belong to the GT5 family and appear to lack these regulatory features, although both the GT3 and GT5 families have a GT-B architecture comprised of two tightly associated Rossmann fold domains (13, 17). GS, unlike GN, lacks the ability to initiate glucose chain formation and instead only catalyzes the extension of primed chains generated by GN. Because GN is a constitutively active enzyme, glycogen synthesis is regulated at the level of GS catalytic function. GS activity is tightly repressed by phosphorylation (1, 18) on regulatory sites within N-terminal (sites 2 and 2a) and C-terminal (sites 3a, 3b, and 3c) extensions to the core Rossmann fold domains that differ significantly in sequence between metazoan and fungal species. GS activity is potently activated by the combination of dephosphorylation and the binding of the allosteric activator glucose-6-phosphate (G6P) (1, 19, 20). The structures of the yeast GS enzyme have yielded insights into allosteric regulation by G6P and provide a template for understanding phosphoregulation in fungal species, features of which are likely conserved in metazoan orthologs (19). The structural basis by which GS and GN interact and how this interaction contributes to glycogen synthesis remains uncharacterized.
Here, we report the X-ray crystal structure of GS in complex with a minimal targeting region of GN from C. elegans. The structure reveals that CeGN binds a conserved surface on CeGS that is remote from previously characterized sites for G6P, UDP, sugar, and tetramer interactions. This interaction surface is required for glycogen synthesis in vitro and in vivo, and the CeGS–CeGN interaction is enhanced by an avidity effect between the CeGN dimer and the CeGS tetramer. Finally, the structure also reveals conserved features of the phosphoregulatory elements of CeGS. Collectively, these results explain how CeGN initiates glycogen chain synthesis by CeGS.
Results and Discussion
Determination of a CeGS–CeGN Costructure.
We surveyed a number of GS–GN pairs from metazoan species, including Homo sapiens, Xenopus laevis, Drosophila melanogaster, Danio rerio, and C. elegans, for high level expression in bacteria. Full-length CeGS and CeGN were both well expressed and chosen as a model to understand the GS–GN interaction and GS regulation. Gene names and accession numbers for GS and GN proteins are provided in SI Appendix, Table S1.
Residues 301–333 of the C-terminal tail of HsGN1 are sufficient for interaction with GS (15) and this sequence is well conserved across metazoan species (SI Appendix, Fig. S1). A fragment that encompassed residues 268–302 of CeGN denoted CeGN34 (Fig. 1A) bound to CeGS with a Kd of 1.6 ± 0.3 μM (Fig. 1B). Although we pursued protein crystallization trials of CeGS in complex with full-length CeGN or CeGN34, only the complex of full-length CeGS (residues 1–672) in complex with CeGN34 yielded crystals, which diffracted to better than 2.7 Å resolution. The structure was solved by molecular replacement using yeast GS coordinates as a search model (19). Following manual rebuilding and refinement of four CeGS protomers in the asymmetric unit, electron density corresponding to four copies of CeGN34 was clearly apparent (SI Appendix, Fig. S2A). Manual building of the CeGN34 sequence and subsequent refinement yielded a final model with good crystallographic and geometric statistics (Rwork/Rfree = 0.179/0.224; see SI Appendix, Table S2 for data collection and refinement statistics).
The refined structure consisted of a tetramer of CeGS with each protomer engaging a single protomer of CeGN34 at its outer periphery (Fig. 1C). Crystal packing analysis revealed this interaction as the only common interface between CeGN34 and each CeGS protomer (SI Appendix, Fig. S3). The tetrameric arrangement of CeGS protomers closely resembled the tetrameric arrangement of ScGS (also known as Gsy2p) in the absence of G6P (rmsd = 2.0 Å versus 2.5 Å for the yeast ScGS tetramer bound to G6P) (19). Each CeGS protomer displayed a closed conformation of the active site, which arises from close interprotomer contacts (SI Appendix, Fig. S4) and confers low basal enzyme activity (19). The binding surface of CeGN34 on CeGS was evolutionarily conserved but nonoverlapping with previously identified ligand binding sites for UDP-G, G6P, and α1,4-linked glucose (SI Appendix, Fig. S5). The CeGN binding surface was also distinct from the intramolecular binding sites for the N- and C-terminal regulatory tails in the previously characterized yeast structures (19), and in the C. elegans structure as described below (SI Appendix, Fig. S5).
The GS Phosphoregulatory Apparatus and Implications for Regulation.
The structure of the individual CeGS protomers was highly similar to that of the ScGS protomers (20) with the important exception that our structure contained both phosphoregulatory regions of metazoan GS, namely the extended N-terminal tail that is absent in yeast ScGS and the C-terminal tail, which was partially disordered in the ScGS structure (Fig. 1D and SI Appendix, Fig. S6–S8). The N-terminal segment consisted of a short loop L1 (residues 7–12), a short helix α1′ (residues 13–21), a long meandering loop L2 (residues 22–36), and finally a short helix α2′ (residues 37–42) (SI Appendix, Fig. S6A). Helix α1′, which houses phosphoregulatory sites 2 and 2a (Ser12 and Thr19), was situated at the center of a globular motif that packed against a conserved exposed surface of CeGS at the junction of the two Rossmann fold domains (SI Appendix, Fig. S6 A and B). Tethering interactions included hydrophobic contacts across Met7, Pro8, Leu11, Ile16, Ile20, Leu25, Leu31, and Met33 on the regulatory tail with Ile70, Val624, and Phe625 on Rossmann fold domain 1 and His562, Val563, Leu613, and Leu616 on Rossmann fold domain 2. Notable charge complementary interactions within the globular motif were observed between Lys15 and Lys18 with Glu23, Asp26, and Glu29. For comparison, the corresponding N-terminal tail sequence of yeast GS is only six residues in length and lacks phosphoregulatory sites (SI Appendix, Fig. S7).
The C-terminal phosphoregulatory tail consisted of a single helix α24 followed by a 16-residue meandering loop (Fig. 1D and SI Appendix, Fig. S6A) that harbors phosphoregulatory sites 3a, 3b, and 3c (Ser654, Ser658, and Ser662, respectively). The position of helix α24 was rotated ∼90° relative to the corresponding helix in yeast GS and, in addition, the following loop region was projected in an opposite direction (Fig. 1D). The C-terminal regulatory tail of CeGS was anchored by hydrophobic interactions involving Val641, Met645, and Val652 in the CeGS C-terminal tail with Tyr189, Thr190, Leu192, and Leu634 on the first Rossmann fold domain (SI Appendix, Fig. S6C). The last seven residues of the C-terminal tail are likely disordered in solution because they were only ordered in one of four CeGS protomers because of crystal packing interactions (SI Appendix, Fig. S9).
The transition between open (active) and closed (inactive) conformations of GS active sites has been hypothesized to be regulated in part by intra- (cis) or inter- (trans) protomer interactions of phosphoregulatory sites with the Arg cluster (Arg600, 601, 603, and 607) at the head of helix α22 (19). Relative to the position of Arg601, sites 2 and 2a are remotely positioned within 23–30 Å in cis and 6–16 Å in trans, whereas sites 3a, 3b, and 3c are positioned within 58–71 Å in cis and 41–54 Å in trans. These separations necessitate a partial unraveling of the phoshoregulatory apparatus to achieve the proposed binding mode. Ser12 and Thr19 at either end of helix α1′ are well positioned to cause dissociation of the N-terminal tail from its ordered position in response to phosphorylation and, thereby, facilitate engagement with the Arg cluster. Sites 3a, 3b, and 3c lie in an intrinsically disordered region of the C-terminal tail and, thus, phosphorylation of these sites is not expected to enhance accessibility to the Arg cluster to a similar degree. However, because helix α24 of the C-terminal regulatory tail is buttressed against helix α2′ of the N-terminal regulatory tail, cumulative phosphorylation of all of the regulatory sites may afford cooperative disengagement of the regulatory regions from their binding sites, as supported by previous mutational analyses (21). By virtue of its binding mode to the hinge region between Rossmann fold domains, dissociation of the N-terminal tail from its intramolecular binding site might also influence enzyme function by affecting interlobe dynamics (17). Lastly, the N-terminal regulatory tail of each protomer participates in interprotomer contacts within the CeGS tetramer (SI Appendix, Fig. S10). Reorganization of the N-terminal tail induced by phosphorylation may therefore impact the closed versus open interprotomer arrangement and, thereby, directly affect enzyme function, in addition to facilitating phosphosite interactions with the Arg cluster.
The GS–GN Interaction Surface.
Analysis of the crystal contacts between CeGN34 and CeGS protomers revealed that the largest total surface area buried by each binary interaction of CeGS with CeGN34 was 2,157 Å2 (SI Appendix, Fig. S3). Each protomer of CeGN34 engaged the first of the two Rossmann fold domains of a CeGS protomer by using identical binding modes (rmsd = 0.7 Å) across all four CeGS–GN34 complexes in the asymmetric unit (SI Appendix, Fig. S2B). The structure of CeGN34 consisted of two α-helices, denoted αBH1 and αBH2, joined by an 11 residue meandering linker (Fig. 1C). This helix-turn-helix motif engaged a complementary “V” shaped pocket on CeGS (Fig. 1 D and E), comprised of helices α4, α6, α9, and α10 (Fig. 1E). Contacts of CeGS with CeGN34 were mediated by a mixture of hydrophobic and hydrogen-bonding interactions (see Fig. 1E for a full listing of contact residues and interactions).
Notable hydrophobic interactions involving αBH1 and the following turn motif of CeGN34 involved CeGN residues Thr270, Arg273, Arg274, Trp277, Pro282, and Tyr284 with CeGS residues Phe151, Ile156, Pro159, Cys261, and Tyr257 (Fig. 1F). αBH2 of GN was amphipathic in nature, presenting a hydrophobic surface composed by Phe290, Ile293, Leu297, and Leu301 toward a complimentary surface on CeGS composed by Arg264, Cys261, Cys154, Lys155, Arg210, Leu211, and Thr269 (Fig. 1F). Notable hydrogen-bonding interactions across the interaction surface include between Gln268GS and Asn298GN and between Lys255GS, Asp248GS, and Tyr257GS side chains and CeGN34 main chain atoms (Fig. 1F).
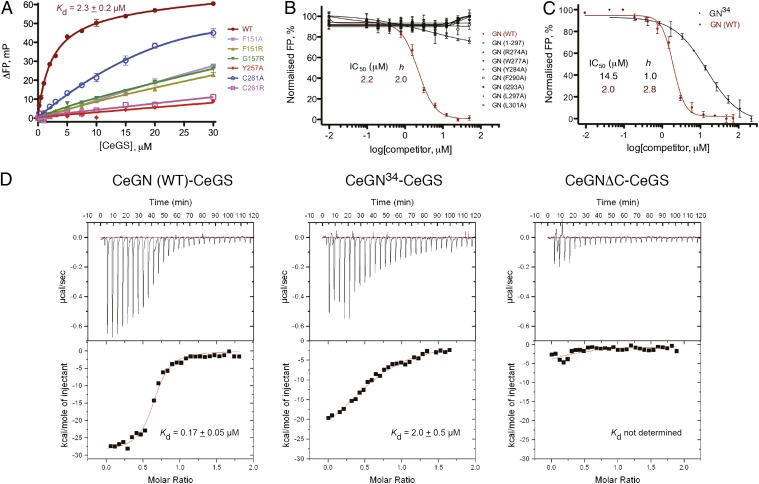
To validate the CeGS–GN34 interaction surface revealed by the X-ray crystal structure, we mutated key contact residues and tested for effects on binding by using a fluorescence polarization (FP) assay (Fig. 2A). We first mutated residues in CeGS and examined binding to wild-type CeGN34 peptide. As a point of reference, wild-type CeGS bound to a fluorescein-labeled CeGN34 peptide with a Kd of 2.3 ± 0.2 µM (Fig. 2A). The substitutions Tyr257Ala and Cys261Arg in CeGS were predicted to disrupt interaction of Rossmann fold domain 1 with all three structural elements of CeGN34 and, indeed, severely perturbed binding (Fig. 2A). The substitutions Phe151Ala, Phe151Arg, and Gly157Arg in CeGS, which were at positions that primarily contacted helix αBH1 of CeGN, had less severe effects, whereas a conservative Cys261Ala mutation had the weakest effect on the CeGS–CeGN interaction (Fig. 2A). All of these effects were consistent with the CeGS–GN34 costructure.
Fig. 2.
Mutational analysis of the CeGS–CeGN34 interaction in vitro. (A) Analysis of binding between CeGN34 and the indicated mutants of CeGS using an FP displacement assay. Results ± SEM are the average of two independent experiments carried out in duplicate. (B) Displacement of fluorescein-labeled CeGN34 tracer peptide by wild-type and mutant CeGN proteins. Results ± SEM are the average of three independent experiments carried out in duplicate. (C) Displacement of fluorescein-labeled CeGN34 tracer by unlabeled CeGN34 peptide and full-length CeGN. h = Hill coefficient. Results ± SEM are the average of three independent experiments carried out in duplicate. (D) Isothermal titration calorimetry analyses for binding of CeGS to full-length CeGN, CeGN34 peptide, and CeGNΔC (residues 1–267). Fitted Kd ± SD values are the mean of two independent experiments, for which a representative trace is shown.
We then mutated the CeGS contact surface on CeGN in the context of full-length CeGN and assessed the ability of mutant proteins to bind full-length CeGS, using an FP-based displacement assay. The full-length CeGN protein displaced a fluorescein-labeled wild-type CeGN34 tracer peptide with an IC50 of 1.9 µM (Fig. 2B), whereas a C-terminal deletion mutant that lacks the entire 34-residue GS-interacting region (CeGN1-297) was inert in this assay (Fig. 2B). Consistent with the X-ray structure, the CeGN mutations Arg274Ala, Trp277Ala, Tyr284Ala, Phe290Ala, Ile293Ala, Leu297Ala, and Leu301Ala all virtually eliminated the interaction with CeGS (Fig. 2B). Together, these results demonstrate that the binding surface observed in the costructure is essential for the high-affinity interaction of CeGS to CeGN in vitro.
Oligomerization of CeGN and CeGS Enhances Binding Through an Avidity Effect.
To assess whether the 34-residue motif of CeGN is the sole determinant for the interaction with CeGS, we compared the ability of CeGN34 and full-length CeGN to bind CeGS in the FP displacement assay. Unlabeled CeGN34 peptide displaced fluorescein-labeled CeGN34 tracer peptide with an IC50 value of 14.5 µM and a Hill coefficient (h) of 1 (Fig. 2C), approximately sevenfold weaker than full-length CeGN (IC50 = 2.0 µM, h = 2.8). A similar trend was obtained by using isothermal titration calorimetry (ITC), with a Kd of 0.17 µM for the CeGS–CeGN interaction compared with a Kd of 2 µM for the CeGS–GN34 interaction (Fig. 2D). The globular domain of CeGN alone did not detectably interact with CeGS (Fig. 2D). To determine whether the poorly conserved region that links the globular domain of CeGN to the CeGN34 motif (SI Appendix, Fig. S1) contributed to the CeGS interaction, we generated a series of CeGN constructs with progressively shortened or lengthened linkers (SI Appendix, Fig. S11A), but we observed no discernible role for the linker region in the CeGS–CeGN interaction (SI Appendix, Fig. S11B). To rule out a potential contribution of covalently attached sugars on full-length CeGN to the interaction with CeGS, we tested a Tyr194Phe CeGN mutant that was incapable of autoglucosylation (SI Appendix, Fig. S12). Glucosylated and nonglucosylated CeGN were equally effective at binding CeGS (SI Appendix, Fig. S12A), thereby ruling out a role for the glycan moiety in the GS–GN interaction, consistent with previous findings (15).
GN and GS are capable of forming dimers and tetramers (12, 19), although GN can function as a monomer at low enzyme concentrations (22). Given the potential for multimer formation, the differences in affinities and Hill coefficients for full-length CeGN versus CeGN34 binding to CeGS could in part be due to an avidity affect. The distance between the C termini of the globular portion of GN protomers within the dimer of rabbit GN structure [Protein Data Bank (PDB) ID code 1ZCU] is ∼94 Å (SI Appendix, Fig. S13), whereas the three unique distances between the N-termini of bound CeGN34 protomers in our structure were ∼144 Å (chains A and B), ∼104 Å (chains B and C), and ∼141 Å (chains A and C). These separations could, in principle, enable three different binding modes (SI Appendix, Fig. S13). In the absence of rearrangement to the GS structure, GN proteins with short linkers may have a preference for engaging GS through the shortest distance (chains A to D and chains B to C with equal distance), whereas for GN proteins with long linkers, engagement of GS through all three distances might be possible. Interestingly, only two of the three interaction modes (chains A to B and chains B to C) provide a direct line of sight between the active sites of GN and GS that would be expected to more easily support glycogen chain elongation (SI Appendix, Fig. S13).
The GS Interaction Motif of GN Is Required for Glycogen Production in Vitro.
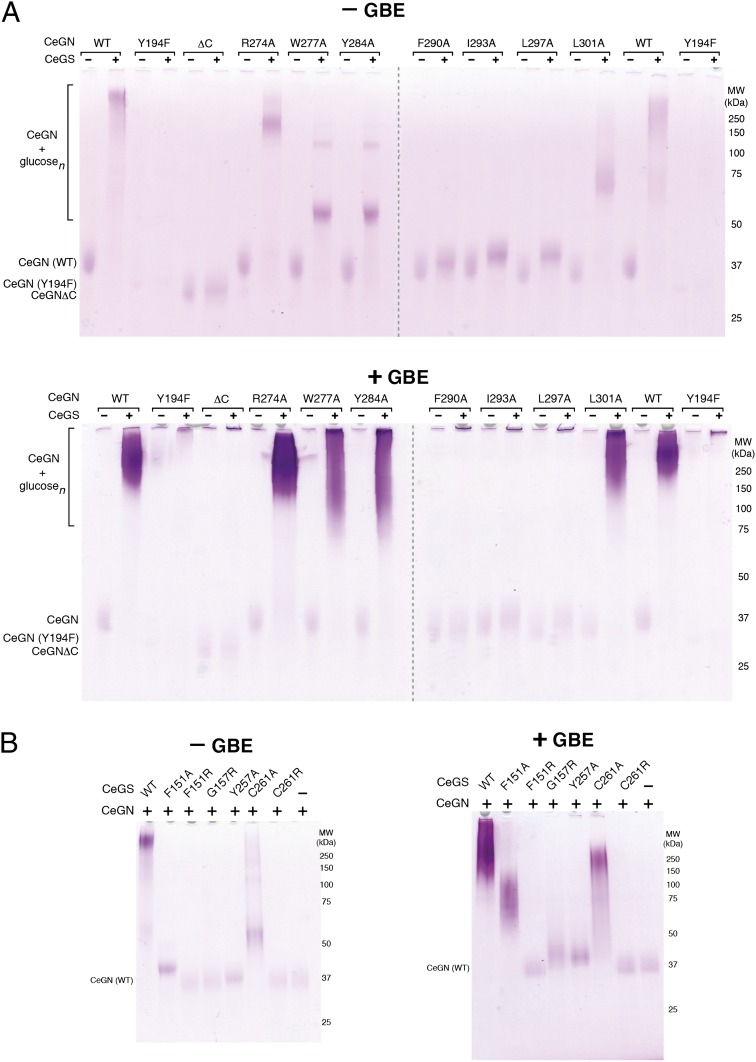
After establishing the structural determinants for the high-affinity interaction of CeGS with CeGN in solution, we investigated the effects of binding interface mutations on the ability of CeGS to produce glycogen de novo on self-primed CeGN produced in bacteria (SI Appendix, Fig. S12B). We used a gel-based assay to monitor glycogen particle size in the presence or absence of G6P and in the presence or absence of glycogen branching enzyme (GBE) (SI Appendix, Fig. S14). Glycogen was detected by a periodic acid-Schiff stain (PAS) or a Coomassie dye stain. Wild-type GN served as an excellent substrate for glucosylation by CeGS as evidenced by the production of large molecular mass species (Fig. 3A). In contrast, no glucosylated species were detected with either a catalytically inactive CeGNTyr194Phe mutant or the CeGNΔC mutant that is defective for interaction with CeGS (Fig. 3A). The CeGN mutants Phe290Ala, Ile293Ala, and Leu297Ala were also severely compromised glucosylation substrates for CeGS (Fig. 3A), whereas the Arg274Ala, Trp277Ala, Tyr284Ala, and Leu301Ala mutants displayed intermediate abilities to serve as substrates (Fig. 3A). When performed in the presence of GBE, which greatly increases the number of reducing ends available for extension, the CeGN mutants Phe290Ala, Ile293Ala, and Leu297Ala remained fully compromised for function (Fig. 3A), whereas the Arg274Ala, Trp277Ala, and Leu301Ala mutants approached the activity of wild-type CeGN in the endpoint assay (Fig. 3A).
Fig. 3.
The CeGS–CeGN34 interaction is required for glycogen formation. (A) Glucosylation of bacterially expressed wild-type and mutant forms of CeGN by CeGS. Reactions were performed in the absence and presence of GBE. The CeGN Y194F mutant does not self-glucosylate and is a negative control. SDS/PAGE gels were visualized by periodic acid–Schiff staining. Results are representative of three independent experiments. (B) Glucosylation of bacterially expressed CeGN by wild-type and mutant forms of CeGS. Experiments were performed as in A.
Reciprocal surface mutations on CeGS caused similar effects (Fig. 3B). In the absence of GBE, the CeGS mutants Phe151Arg, Gly157Arg, Tyr257Ala, and Cys261Arg were fully compromised for the ability to extend glucosyl chains on CeGN, whereas the CeGS mutants Phe151Ala and Cys261Ala retained some activity (Fig. 3B). Addition of GBE partially overcame the defect in the latter reactions (Fig. 3B). Deletions and insertions within the linker of CeGN that had no effect on the CeGS interaction also had no effect on particle size distribution for enzymatic reactions carried out in the absence of GBE (SI Appendix, Fig. S11 C, Left), but caused modest effects for reactions carried out in the presence of GBE (SI Appendix, Fig. S11 C, Right). These latter effects displayed an interesting trend in that shorter linkers resulted in glycogen species of smaller maximal size, an increased mean size, and a narrower overall size distribution, whereas longer linkers caused the opposite effects (SI Appendix, Fig. S11C). Together, these results validated the functional importance of the CeGS–CeGN interaction and suggested a possible role for CeGN linker length in setting glycogen particle size.
The GS–GN Interaction Is Required for Glycogen Accumulation in Yeast.
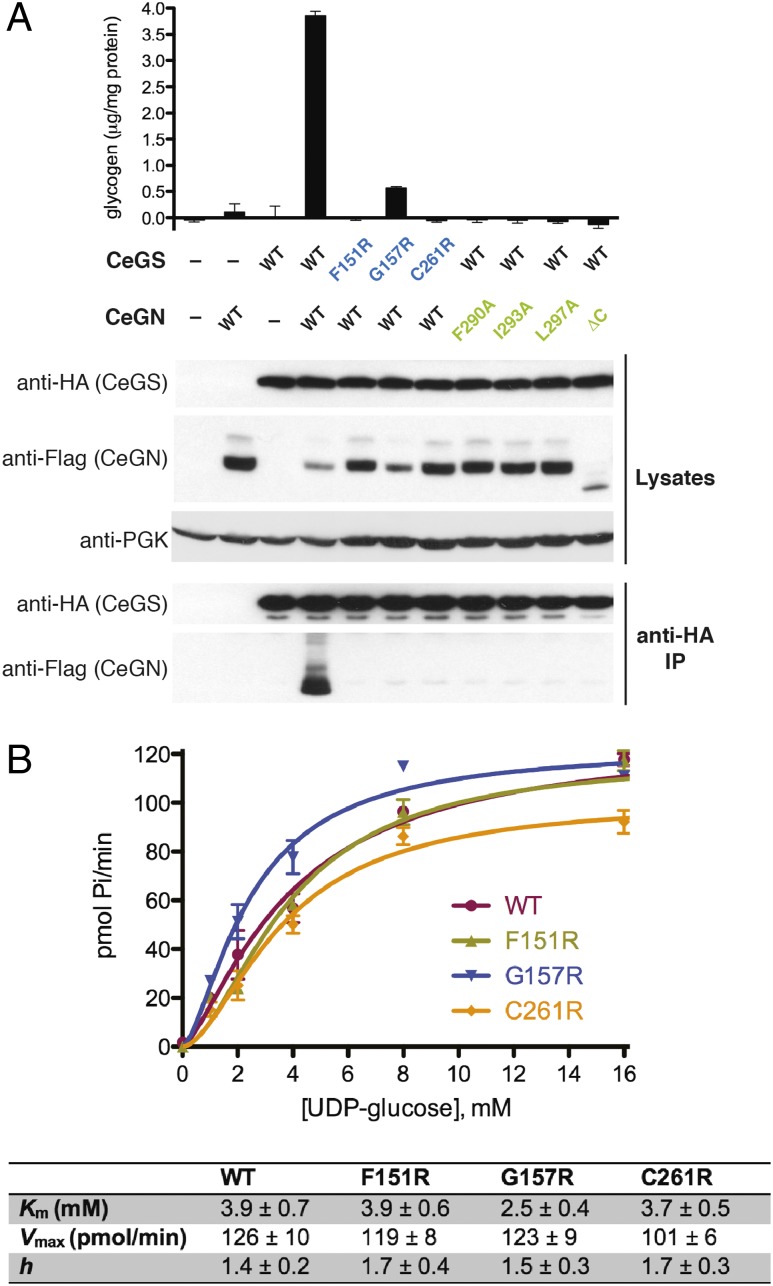
To ascertain the role of the GS–GN interaction in vivo, we expressed wild-type and mutant forms of CeGS and CeGN proteins in a quadruple mutant yeast strain that is deleted for each of CeGS and CeGN genes, i.e., a gsy1Δ gsy2Δ glg1Δ glg2Δ strain. As expected, the quadruple deletion mutant did not accumulate glycogen and introduction of either CeGS or CeGN alone did not rescue this defect (Fig. 4A). However, when CeGS and CeGN proteins were expressed together from a constitutive promoter, glycogen production was restored. When expressed with CeGN, the CeGS mutants Phe151Arg and Cys261Arg failed to restore glycogen production, whereas the CeGS mutant Gly157Arg displayed a residual level of glycogen accumulation (Fig. 4A). Similarly, when expressed with CeGS, the CeGN single-site mutants Phe290Ala, Ile293Ala, Leu297Ala, and the deletion mutant CeGNΔC also failed to restore glycogen production (Fig. 4A). The mutant proteins were expressed at comparable levels and were compromised for CeGS–CeGN interactions in vivo, (Fig. 4A), as expected from in vitro data (Fig. 3). Importantly, the intrinsic enzymatic activity of each CeGS interaction mutant, as measured by Km and Vmax for elongation of free maltooctaose chains in the presence of GBE, were indistinguishable from wild-type CeGS (Fig. 4B), demonstrating that the glycogen accumulation defect in vivo was due to the loss of the CeGS–CeGN interaction. These results demonstrate that the CeGS–CeGN interaction mediated by the CeGN34 motif is essential for glycogen accumulation in a yeast surrogate model.
Fig. 4.
The CeGS–CeGN34 interaction is required for glycogen formation in yeast. (A) The indicated versions of CeGS and CeGN were expressed in a gsy1Δ gsy2Δ glg1Δ glg2Δ quadruple deletion yeast strain that lacked endogenous GS and GN. After cell lysis, samples were analyzed for glycogen accumulation (Upper) and for GS–GN interaction by anti-HA immunoprecipitation and either an-HA or anti-FLAG immunoblot (Lower). Anti-PGK (phosphoglycerate kinase) signal was used as a loading control. (B) Glucosylation activity of wild-type and mutant forms of GS expressed and purified from the gsy1Δ gsy2Δ glg1Δ glg2Δ yeast strain. The ability of GS to extend maltooctaose chains in the presence of GBE, 3 mM G6P and increasing UDP-G concentrations was assessed by monitoring phosphate (Pi) release in a coupled colorimetric assay. Kinetic parameters for UDP-G ± SD are the average of two independent experiments.
Role of the GS–GN Interaction in Mouse Cells.
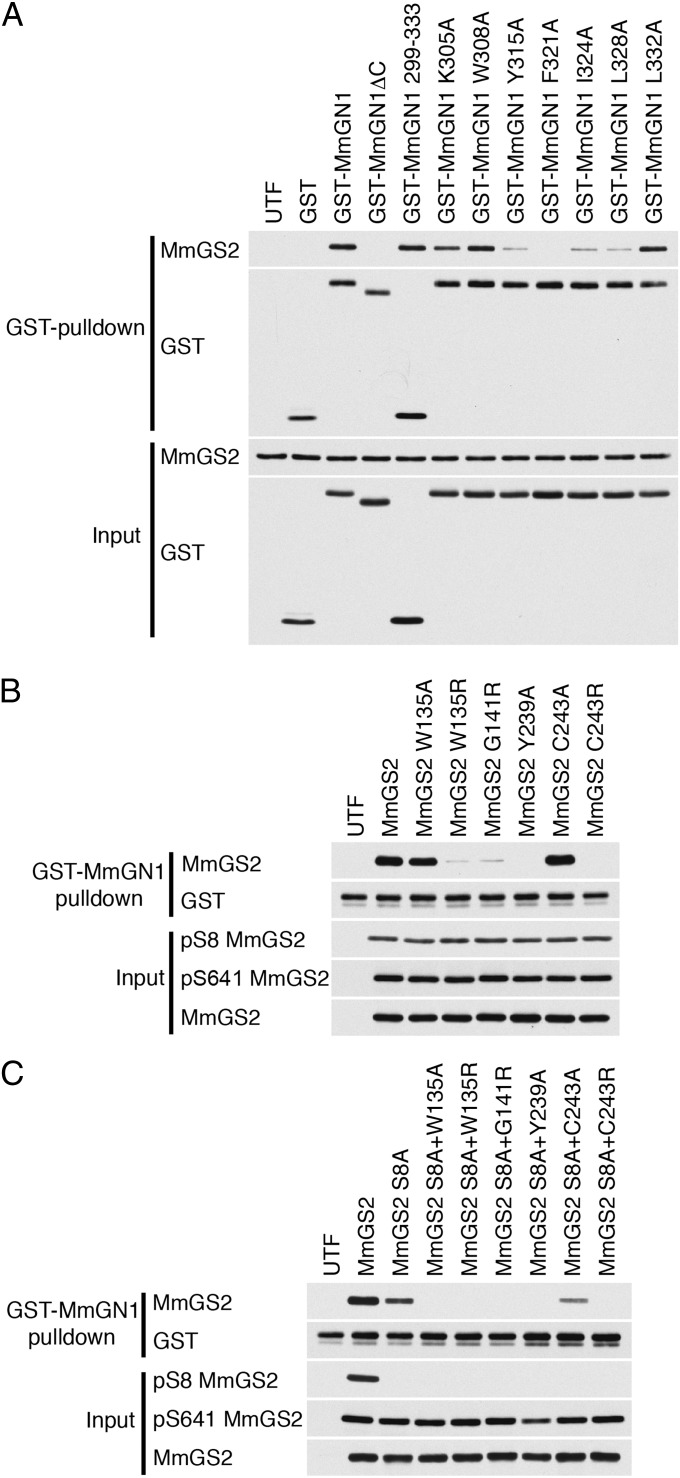
We next investigated the role of the GS–GN interaction in a mammalian model system. The mouse genome encodes a single copy of GN (MmGN1) and two copies of GS (MmGS1 and MmGS2; SI Appendix, Table S1). Using the CeGS-GN34 structure as a guide, we mutated key residues predicted to mediate the MmGN1–MmGS2 interaction (SI Appendix, Figs. S1 and S7). Coexpression of wild-type and mutant MmGN1 and MmGS2 by transient transfection in COS1 cells revealed variable levels of protein expression. For this reason, we expressed GST-tagged MmGN1 and untagged MmGS2 individually and mixed cell lysates to achieve comparable protein levels before the isolation of stable complexes by affinity capture with glutathione resin (Fig. 5A). As expected, MmGS2 and the 35-residue C-terminal region alone (MmGN1299-333) bound robustly to MmGS2, whereas a deletion mutant MmGN1ΔC that lacked the 35 C-terminal residues was unable to bind MmGS2 (Fig. 5A). The MmGN1 point mutants Tyr315Ala, Phe321Ala, Ile324Ala, and Leu328Ala (analogous to the strong CeGN mutants Tyr284Ala, Phe290Ala, Ile293Ala, and Leu297Ala) exhibited a drastic reduction in the MmGS2 interaction, whereas the MmGN1 point mutants Lys305Ala, Trp308Ala, and Leu332Ala (analogous to the weak CeGN mutants Arg274Ala, Trp277Ala, and Leu301Ala) retained residual binding to MmGS2 (Fig. 5A). Similarly, in a capture assay with bacterially expressed GST-MmGN1, the MmGS2 point mutants Trp135Arg, Gly141Arg, Tyr239Ala, and Cys243Arg (analogous to the strong CeGS mutants, Phe151Arg, Gly157Arg, Tyr257Ala, and Cys261Arg) all exhibited reduced binding to MmGN1 (Fig. 5B), whereas the MmGS2 mutants Trp135Ala and Cys243Ala (analogous to the weak CeGS mutants Phe151Ala and Cys261Ala) retained appreciable binding to MmGS2 (Fig. 5B). The effect of each MmGS2 mutation displayed the same general trend when combined with a mutation of phosphoregulatory site 2 (Ser8Ala) (Fig. 5C).
Fig. 5.
The GS–GN34 interaction in mammalian cells. (A) COS1 cells were transfected with the indicated GST-MmGN1 constructs in pEBG6P or MmGS2 (untagged) constructs in pCMV5. GST-MmGN1 and MmGS2 transfected lysates were mixed, and complexes were captured on glutathione resin and analyzed by immunoblotting with the indicated antibodies. Results are representative of three independent experiments. UTF, untransfected. (B) COS1 cells were transfected with the indicated MmGS2 constructs in pCMV5 vector. MmGS2 was captured with bacterial GST-MmGN1 on glutathione resin and immunoblotted with the indicated antibodies. Input lysates were adjusted for differences in MmGS2 expression. Results are representative of three independent experiments. UTF, untransfected. (C) As in B, except that each GS mutation was combined with a Ser8Ala phosphorylation site mutation.
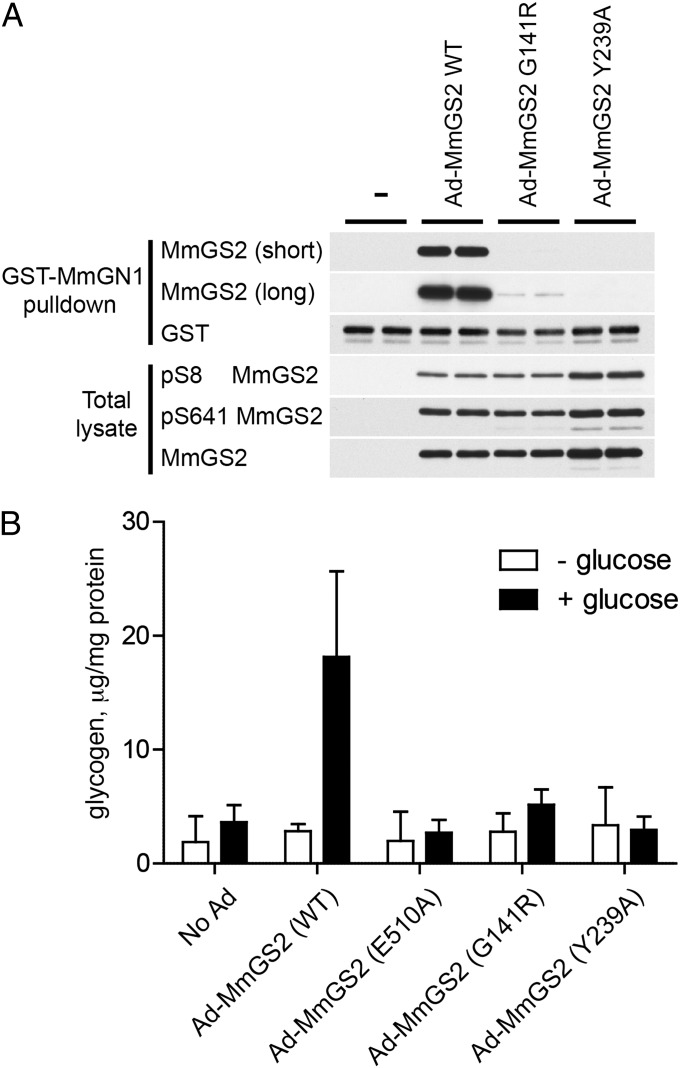
To investigate the effect of the murine GS–GN interaction on glycogen accumulation function in intact cells, we used cultured hepatocytes isolated from an MmGS2 knockout mouse model (23). We used an adenovirus system to infect MmGS2-deficient hepatocytes with either wild-type or mutant forms of MmGS2. We used a GST-MmGN1 fusion protein expressed in bacteria to capture adenovirus-expressed untagged MmGS2 proteins on glutathione resin. As expected, wild-type MmGS2, but neither the Tyr239Ala nor the Gly141Arg mutant form, was captured on the MmGN1 resin (Fig. 6A). Measurement of glycogen content revealed that cells expressing the MmGS2 Gly141Arg or Tyr239Ala mutants, or a Glu510Ala catalytically inactive mutant as a negative control, failed to accumulate glycogen in the presence of a high level (25 mM) of glucose, in contrast to cells that expressed wild-type MmGS2 (Fig. 6B). Importantly, phosphorylation levels of MmGS2 at sites 2 (Ser8) and 3a (Ser641) were similar between the wild-type and mutant MmGS2 forms (Figs. 5 B and C and 6A), thereby ruling out any overt defect in phosphoregulation. We ruled out major intrinsic defects of the mutant MmGS2 enzymes through measurement of the Km for UDP glucose and the Ka for G6P for each of the mutant and wild-type enzymes (SI Appendix, Fig. S15). We also ascertained that levels of the known modulators of glycogen accumulation, including glycogen phosphorylase (GP), phospho-S15 GP, and glucokinase (GCK) proteins, were comparable in each infected cell line (SI Appendix, Fig. S15). The loss of glycogen accumulation in the mutant MmGS2 cells is thus directly attributable to loss of the GS–GN interaction. These results demonstrate the conserved role of the GS–GN interaction in mammalian cells.
Fig. 6.
The MmGS2–MmGN1 interaction is required for glycogen synthesis in mouse primary hepatocytes. (A) Hepatocytes were isolated from MmGS2−/− mice and infected with adenovirus (Ad) encoding MmGS2 wild-type (WT) or the indicated MmGS2 mutants followed by overnight incubation with glucose in FBS-free DMEM. GS was captured on GST-MmGN1 and analyzed by immunoblot with the indicated antibodies (short, short exposure; long, long exposure). Results are representative of two independent experiments. (B) MmGS2−/− hepatocytes were treated and infected with the constructs as described in A. After overnight incubation in FBS-free DMEM, cells were incubated with or without 25 mM glucose for a further 6 h, followed by determination of glycogen levels. A catalytically inactive MmGS2 mutant (Glu510Ala) was used as control. Results ± SD are the average of three independent experiments.
Conclusion
The synthesis of glycogen is a critically important biological process that is used by all eukaryotes for storage of energy in a readily accessible form. Glycogen synthase enzyme activity is tightly regulated through the action of G6P as an allosteric activator, through multiple inhibitory phosphorylation events, and through its interactions with glycogenin. Our structure of the CeGS–CeGN complex demonstrates the basis for recruitment of GS to the GN initiation complex, and we show that this interaction is essential for glycogen synthesis in vitro and in vivo. Our structural data provide a framework for understanding the complex effects of phosphorylation on the interactions of the N- and C-terminal regulatory regions of GS with the catalytic domain. Our data further suggests that the length of glycogen chains synthesized by GS may, in part, be determined by the length of the linker sequence between the catalytic domain and GS-binding domain of GN. This linker exhibits considerable variability through evolution, and we conjecture that linker length may confer subtly different characteristics on glycogen particles in different tissues and across species. The structural understanding of the basis for the GS–GN interaction and GS regulation may allow the development of novel pharmacological modulators of GS activity, with potential therapeutic applications in disorders of glucose metabolism, including glycogen storage disease and diabetes.
Materials and Methods
Cloning, Protein Expression, and Purification of C. elegans GS.
Full-length CeGS was cloned into a pProEx-HTa expression vector by using SfoI/NotI restriction sites. N-terminally 6-His–tagged CeGS was expressed in Escherichia coli BL21 (DE3) cells. Cells were grown in TB medium to A600 = 2 at 37 °C, before protein expression was induced by the addition of 250 µM isopropyl-β-d-thiogalactopyranoside and incubated for an additional 16 h at 20 °C. Cells were harvested by centrifugation for 30 min at 3,500 × g and resuspended in ice-cold lysis buffer. Cells were lysed by using an Avestin C3 cell disrupter (18,000 psi). Lysate was cleared by centrifugation at 30,000 × g for 30 min and filtration by using a 0.45-µm filter before loading onto a 5-mL HiTrap IMAC HP column (GE Healthcare). Following washing with high salt wash buffer B, GS was eluted with a gradient of 20–300 mM imidazole. Fractions containing GS were pooled, and 0.3–0.5 mg of His-tagged tobacco etch virus (TEV) protease was added before dialysis against 4 L of wash buffer A (containing 50 mM NaCl) for 12–14 h. The TEV protease and uncleaved GS were removed by subtraction on HiTrap IMAC resin, then loading on a 5-mL HiTrap-Q column (GE Healthcare). Bound GS was eluted by using a salt gradient of 0–600 mM NaCl. Eluted fractions containing GS were pooled, concentrated, and finally resolved on a Sepharose-S200 16/60 column (GE Healthcare), preequilibrated in 25 mM Tris⋅HCl at pH 7.6, 150 mM NaCl, and 1 mM tris(2-carboxyethyl)phosphine (TCEP). Eluted peaks were analyzed by SDS/PAGE, and fractions containing >95% pure CeGS were combined, concentrated to 5–7 mg/mL, snap frozen in liquid nitrogen, and stored at −80 °C.
Peptide Synthesis.
Unlabeled C. elegans GN C-terminal peptide (GN34) with amino acid sequence (PSTEERRAAWEAGQPDYLGRDAFVHIQEALNRALN) comprising residues 268–302 was synthesized (>95% purity; GL Biochem [Shanghai] Ltd) and used for crystallography and GS-GN34 binding studies (note that while the peptide was 35 residues in length, we refer to this as CeGN34 since the last residue was disordered in the crystal structure). For fluorescence polarization assays, a peptide with the same sequence was synthesized (>95% purity; Biomatik), and used as a tracer by conjugating 5(6)-carboxyfluorescein through a β-alanine linker at the peptide N terminus. Peptides were dissolved in 25 mM Hepes-NaOH at pH 7.5 at a concentration of 0.5–1 mM, aliquoted, and stored at −20 °C. Concentration of the fluorophore linked peptide was calculated by using the Beer–Lambert law by measuring the absorbance at 492 nm and an extinction coefficient of 83,000 M−1⋅cm−1. Concentration of the unlabeled peptide was calculated by the same method by measuring the absorbance at 280 nm and using an extinction coefficient of 6,990 M−1⋅cm−1.
Crystallization, Structure Solution, and Refinement.
CeGS at 52 µM was mixed with 80 µM CeGN34 synthetic peptide (CeGN residues 268–302) and allowed to equilibrate for 30 min on ice. Crystals were grown at 20 °C in sitting drops by mixing 1 µL of protein–peptide complex with 1 µL of mother liquor consisting of 100 mM bis-Tris propane (pH 7.25), 200 mM NaSO4, and 22% (wt/vol) PEG 3350. Needle-shaped crystals appeared overnight and grew to full size over 72 h. Crystals were flash frozen in liquid nitrogen by using mother liquor supplemented with 22.5% (vol/vol) glycerol as cryoprotectant. Data were collected at 100 K on station 24-ID-C, NE CAT beamline, Advanced Photon Source, and processed by using the program XDS (24) (SI Appendix, Table S2). The structure was solved by molecular replacement using the program PHASER (25) and the structure of a single protomer of ScGS (PDB ID code 3NAZ; 19) as a search model. The structure was refined by iterative rounds of refinement with REFMAC (26) and PHENIX (27) and manual model building with the program COOT (28).
Structure Analysis and Sequence Alignments.
Multiple sequence alignments were performed using MUSCLE (29) and displayed, edited and annotated by using ALINE (30). Secondary structure was analyzed by using DSSP (31), and buried surface area and residue contacts were calculated using the programs AREAIMOL and CONTACT from the CCP4 suite (32). Structure alignments and structure representations were performed using The PyMOL Molecular Graphics System, Schrödinger, LLC.
Fluorescence Polarization Peptide Binding Assays.
Binding reactions were performed with 2.5 nM fluorophore-conjugated peptide and the indicated protein concentrations in FP buffer consisting of 50 mM Hepes-NaOH at pH 7.5, 100 mM NaCl, 1 mM TCEP, 0.03% Brij-35, and 1 mg/mL BSA. Binding reactions were allowed to equilibrate for 90 min in 20-μL reactions in 384-well black flat-bottom low flange plates (Corning; 35373). Fluorescence intensities were read by an Analyst HT fluorimeter (Molecular Devices; excitation filter: 485 nm, 20 nm bandwidth; emission filter: 530 nm, 25 nm bandwidth; dichroic filter with a cutoff of 505 nm; 10 readings per well; time between readings: 100 ms; integration time: 1 s; motion settling time: 25 ms). Fluorescence polarization was calculated with the LJL Criterion Host Software by using the formula FP (in polarization units, P) = (Fparallel − Fperpendicular)/(Fparallel + Fperpendicular) and a G factor of 0.92 (Fparallel and Fperpendicular, fluorescence intensities parallel and perpendicular to the excitation plane). Kd values were calculated by nonlinear regression analysis of FP values performed in GraphPad Prism 5 by using a one-site specific binding model: Y = Bmax × X/(Kd + X). For competition binding assays, the CeGS concentration was fixed at 3.5 µM (corresponding to ∼80% of total binding signal) in the presence of varying concentrations of competitor. Data were fitted by using the Prism built-in dose–response equation for inhibition with variable slope: Y = Bottom + (Top − Bottom)/(1 + 10((LogIC50−X) × h)).
Isothermal Titration Calorimetry.
Isothermal titration calorimetry was performed at 22 °C by using a MicroCal VP-ITC titration microcalorimeter. CeGS at a concentration of 10–15 μM in 50 mM Hepes-NaOH at pH 7.5, 150 mM NaCl, and 1 mM TCEP was placed in the 1.4-mL calorimeter cell and sevenfold excess titrant in the same buffer was added sequentially in 10-μL aliquots (for a total of 30 injections) at 4-min intervals. The heat of reaction per injection (microcalorie/second) was determined by integration of the peak areas using the Origin Version 5.0 scientific plotting software. Curve fitting was performed by using the same software using a one-site binding model.
Glycogen Extraction and Quantification from Yeast.
A yeast strain deleted for endogenous GS and GN genes (quadruple glg1, glg2, gsy1, and gsy2 knockout) was generated by sequential replacement of glg1, gsy1, and gsy2 genes with cassettes NATMX4, HPHMX4, and HIS3MX6, respectively, in a glg2 deletion strain (glg2::kanMX4; kindly provided by Charlie Boon, University of Toronto, Toronto). The resultant strain was transformed with plasmids expressing CeGS (HA-tagged) or CeGN (Flag-tagged) from ADH promoter. Cells were grown to mid-log phase in selective media, pelleted, and washed in PBS. Cells were then split for protein extraction-immunoprecipitation (see SI Appendix, Immunoprecipitation of CeGS-CeGN from Yeast) and glycogen preparation. For glycogen preparation, cells were lysed in ice-cold PBS by bead beating using Disruptor Genie (Scientific Industries). A small fraction (∼5 μL) of cleared cell lysates was taken for protein quantification, and the remainder was boiled (100 °C) for 5 min to deactivate all proteins. Soluble glycogen was collected in the supernatant after subsequent centrifugation at 15000 × g for 10 min. Four microliters of glycogen sample, equivalent to a volume containing 100 μg of protein were used for glycogen quantification by using a Glycogen Assay Kit (Abcam; ab65620) based on glucoamylase-mediated hydrolysis of glycogen to glucose, which was then reacted with OxiRed to generate color (570 nm) and fluorescence (Ex 540/Em 590) signals. Fluorimetric assays were carried out in 96-well plates (100 μL per well) and read on an Envision 2104 Multilabel Reader (Perkin-Elmer).
GS Activity Assay from Yeast-Expressed Proteins.
HA-tagged wild-type or mutant CeGS proteins were expressed in exponentially growing yeast cells that lack endogenous GS and GN. Proteins were extracted in buffer containing 50 mM Hepes-NaOH (pH 7.5), 150 mM NaCl, 5 mM EDTA, 5 mM NaF, 0.1% Nonidet P-40, 10% glycerol, and supplemented with Complete protease inhibitor mixture (Roche). Proteins were affinity purified with anti-HA conjugated beads (Santa Cruz). Glycosyltransferase activity of 1 μg of GS was assayed in the presence of 1 μM branching enzyme, 6 mM G6P, 10 mM maltooctaose, and 1–24 mM UDP-G in 50 mM Hepes-NaOH at pH 7.5, 150 mM NaCl, and 2 mM EDTA. Reactions were incubated at 30 °C for 30 min before addition of UDP dephosphorylation reagent from Glycosyltransferase Activity Kit (R&D Systems). Dephosphorylation was carried out at 37 °C for 20 min before color development and absorbance reading at 620 nm according to the kit instruction. Kinetic parameters were determined by nonlinear regression using GraphPad Prism and a modified Michaelis–Menten equation incorporating the Hill coefficient: υ = Vmax × [UDP-G]h/(Kmh + [UDP-G]h).
Adenovirus Production and Infection of Hepatocytes.
Generation, purification (cesium chloride banding), and titration of recombinant adenovirus were performed by using the AdEasy system (Agilent Technologies) according to manufacturer’s protocol and the described protocol (33). Primary hepatocytes derived from GS2−/− mice (23) were infected with adenovirus encoding WT MmGS2 and mutants (Glu510Ala, Gly141Arg, Tyr239Ala) by using multiplicity of infection = 8. Precipitation of MmGS2 from Ad-infected hepatocytes was performed as described for COS1 cells. Briefly, ∼5 μg of lysate (normalized by immunoblot) was precipitated with 100 ng of GST-MmGN1 and analyzed by immunoblot as described.
Antibodies used for SI Appendix, Fig. S15 were as follows: pS8 MmGS2 (34), pS641 MmGS2 (CST 3891), total MmGS2 (CST 3886), pS15 GP (sheep polyclonal antibody raised against 10KRKQIpSVRGLA20 from human PYGL by the Division of Signal Transduction Therapy (DSTT) (University of Dundee, Dundee, United Kingdom), S961A, first bleed, GP (sheep polyclonal antibody raised against partially purified rabbit skeletal muscle GP by the DSTT, S956A, second bleed (35), GCK [sheep polyclonal antibody raised against GST-tagged mouse GCK, a generous gift from Mark Magnuson (Vanderbilt University, Nashville, TN)], and GAPDH (Sigma G8795).
Glycogen Extraction and Quantification from Hepatocytes.
The method described by von Wilamowitz-Moellendorff et al. (23) was followed for glycogen extraction. Briefly, cells were scraped into 30% (wt/vol) KOH and the extract was heated for 15 min at 100 °C. Glycogen was selectively precipitated with cold 66% ethanol overnight, and the pellet was digested with α-amyloglucosidase (Sigma). Glucose concentration was determined based on a fluorometric assay described in detail by Saez et al. (36).
Additional experimental procedures are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Daniel Mao and Leanne Wybenga-Groot for help with data collection at the synchrotron and Zhiqin Li for excellent technical support. We thank the Advanced Photon Source on the Northeastern Collaborative Access Team beam lines, supported by National Institute of General Medical Sciences Grant P41 GM103403 from the National Institutes of Health and by the US Department of Energy under Contract DE-AC02-06CH11357. This work was supported by a Human Frontiers Science Program fellowship and a Sir Henry Wellcome postdoctoral fellowship (to E.Z.); Canadian Institutes of Health Research Grant MOP-84370 (to F.S.); British Heart Foundation Grant PG/09/059 and British Medical Research Council (to K.S.); Spanish MINECO Grant BFU2011-30554 (to J.J.G.); Wellcome Trust Grant 085178 and the Ministère de l’Enseignement supérieur, de la Recherche, de la Science et de la Technologie du Québec through Génome Québec (to M.T.); and by Canada Research Chairs in Structural Biology (F.S.) and in Systems and Synthetic Biology (M.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of C. elegans GS–GN34 complex structure have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4QLB).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402926111/-/DCSupplemental.
References
- 1.Roach P. Glycogen and its metabolism. CMM. 2002;2:101–120. doi: 10.2174/1566524024605761. [DOI] [PubMed] [Google Scholar]
- 2.Moslemi AR, et al. Glycogenin-1 deficiency and inactivated priming of glycogen synthesis. N Engl J Med. 2010;362:1203–1210. doi: 10.1056/NEJMoa0900661. [DOI] [PubMed] [Google Scholar]
- 3.Valles-Ortega J, et al. Neurodegeneration and functional impairments associated with glycogen synthase accumulation in a mouse model of Lafora disease. EMBO Mol Med. 2011;3:667–681. doi: 10.1002/emmm.201100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitt Falk Petersen GIS. Cellular mechanism of insulin resistance in skeletal muscle. J R Soc Med. 2002;95:8. [PMC free article] [PubMed] [Google Scholar]
- 5.Ros S, Schulze A. Linking glycogen and senescence in cancer cells. Cell Metab. 2012;16:687–688. doi: 10.1016/j.cmet.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Pitcher J, Smythe C, Campbell DG, Cohen P. Identification of the 38-kDa subunit of rabbit skeletal muscle glycogen synthase as glycogenin. Eur J Biochem. 1987;169:497–502. doi: 10.1111/j.1432-1033.1987.tb13637.x. [DOI] [PubMed] [Google Scholar]
- 7.Pitcher J, Smythe C, Cohen P. Glycogenin is the priming glucosyltransferase required for the initiation of glycogen biogenesis in rabbit skeletal muscle. Eur J Biochem. 1988;176:391–395. doi: 10.1111/j.1432-1033.1988.tb14294.x. [DOI] [PubMed] [Google Scholar]
- 8.Marchand I, et al. Quantification of subcellular glycogen in resting human muscle: Granule size, number, and location. J Appl Physiol. 2002;93:1598–1607. doi: 10.1152/japplphysiol.00585.2001. [DOI] [PubMed] [Google Scholar]
- 9.Roach PJ, DePaoli-Roach AA, Hurley TD, Tagliabracci VS. Glycogen and its metabolism: Some new developments and old themes. Biochem J. 2012;441:763–787. doi: 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker GJ, Koay A, Gilbert-Wilson R, Waddington LJ, Stapleton D. AMP-activated protein kinase does not associate with glycogen α-particles from rat liver. Biochem Biophys Res Commun. 2007;362:811–815. doi: 10.1016/j.bbrc.2007.08.080. [DOI] [PubMed] [Google Scholar]
- 11.Ryu J-H, et al. Comparative structural analyses of purified glycogen particles from rat liver, human skeletal muscle and commercial preparations. Int J Biol Macromol. 2009;45:478–482. doi: 10.1016/j.ijbiomac.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Gibbons BJ, Roach PJ, Hurley TD. Crystal structure of the autocatalytic initiator of glycogen biosynthesis, glycogenin. J Mol Biol. 2002;319:463–477. doi: 10.1016/S0022-2836(02)00305-4. [DOI] [PubMed] [Google Scholar]
- 13.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: Structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 14.Chaikuad A, et al. Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis. Proc Natl Acad Sci USA. 2011;108:21028–21033. doi: 10.1073/pnas.1113921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skurat AV, Dietrich AD, Roach PJ. Interaction between glycogenin and glycogen synthase. Arch Biochem Biophys. 2006;456:93–97. doi: 10.1016/j.abb.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai L, Mu J, Zong H, Depaoli-Roach AA, Roach PJ. Structure and chromosomal localization of the human glycogenin-2 gene GYG2. Gene. 2000;242:229–235. doi: 10.1016/s0378-1119(99)00520-x. [DOI] [PubMed] [Google Scholar]
- 17.Buschiazzo A, et al. Crystal structure of glycogen synthase: Homologous enzymes catalyze glycogen synthesis and degradation. EMBO J. 2004;23:3196–3205. doi: 10.1038/sj.emboj.7600324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palm DC, Rohwer JM, Hofmeyr J-HS. Regulation of glycogen synthase from mammalian skeletal muscle. A unifying view of allosteric and covalent regulation. FEBS J. 2012 doi: 10.1111/febs.12059. [DOI] [PubMed] [Google Scholar]
- 19.Baskaran S, Roach PJ, DePaoli-Roach AA, Hurley TD. Structural basis for glucose-6-phosphate activation of glycogen synthase. Proc Natl Acad Sci USA. 2010;107(41):17563–17568. doi: 10.1073/pnas.1006340107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter RW, Treebak JT, Wojtaszewski JFP, Sakamoto K. Molecular Mechanism by Which AMP-Activated Protein Kinase Activation Promotes Glycogen Accumulation in Muscle. Diabetes. 2011;60(3):766–74. doi: 10.2337/db10-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skurat AV, Wang Y, Roach PJ. Rabbit skeletal muscle glycogen synthase expressed in COS cells. Identification of regulatory phosphorylation sites. J Biol Chem. 1994;269:25534–25542. [PubMed] [Google Scholar]
- 22.Issoglio FM, Carrizo ME, Romero JM, Curtino JA. Mechanisms of monomer and dimer glycogenin autoglucosylation. J Biol Chem. 2011;287(3):1955–61. doi: 10.1074/jbc.M111.287813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilamowitz-Moellendorff von A, et al. Glucose-6-phosphate-mediated activation of liver glycogen synthase plays a key role in hepatic glycogen synthesis. Diabetes. 2013;62:4070–4082. doi: 10.2337/db13-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebedev A, Pannu N, Steiner R. REFMAC5 for the Refinement of Macromolecular Crystal Structures. Acta Crystallogr D Biol Crystallogr 67(4):355–367. [DOI] [PMC free article] [PubMed]
- 27.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Edgar R. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bond CS, Schüttelkopf AW. ALINE: A WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 32.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 33.Luo J, et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 34.Ros S, Garcia-Rocha M, Domínguez J, Ferrer JC, Guinovart JJ. Control of liver glycogen synthase activity and intracellular distribution by phosphorylation. J Biol Chem. 2009;284:6370–6378. doi: 10.1074/jbc.M808576200. [DOI] [PubMed] [Google Scholar]
- 35.Bouskila M, et al. Allosteric regulation of glycogen synthase controls glycogen synthesis in muscle. Cell Metab. 2010;12:456–466. doi: 10.1016/j.cmet.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Saez I, et al. Neurons have an active glycogen metabolism that contributes to tolerance to hypoxia. J Cereb Blood Flow Metab. 2014;34(6):945–955. doi: 10.1038/jcbfm.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.