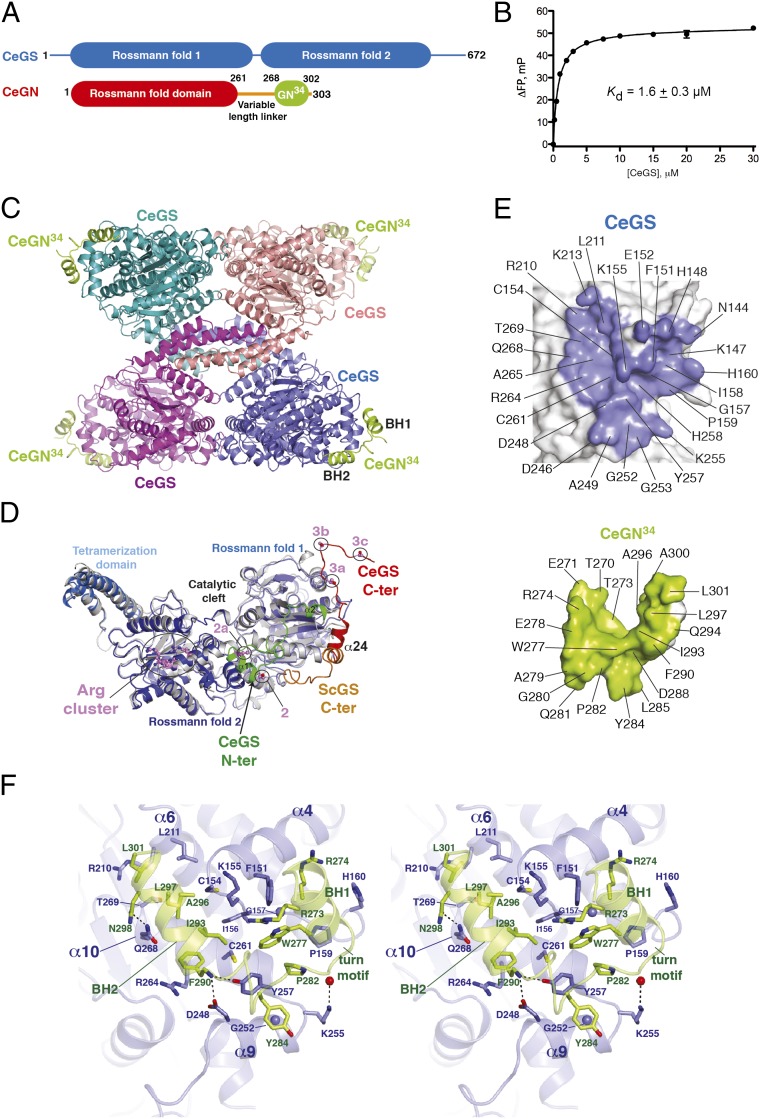
Fig. 1.
Structure of CeGS in complex with CeGN34. (A) Domain architecture of CeGS (GT-B fold) and CeGN (GT-A fold). (B) Interaction of CeGN34 to CeGS determined by fluorescence polarization. Kd value ± SEM is the average of three independent experiments carried out in duplicate. (C) Structure of the CeGS tetramer bound to the CeGN34 peptide. (D) Comparison of CeGS protomers (blue) and the budding yeast ScGS (PDB ID code 3NAZ, gray). The N-terminal extension of CeGS (green) and C-terminal extensions of CeGS (red) and ScGS (orange) are highlighted. Phosphoregulatory sites 2 (S12), 2a (T19), 3a (S654), 3b (S658), and 3c (S662) and the allosteric regulatory site (Arg cluster) on the CeGS protomer are shown as ball and stick models with violet-colored carbon atoms. (E) Peel away surface representations of the CeGS–CeGN complex with contact residues (CeGS, Upper; CeGN, Lower). (F) Detailed stereoview of the CeGS–CeGN34 complex. CeGS is shown in blue and CeGN in green. Side chain residues that make direct contacts are shown as sticks with colored heteroatoms (oxygen, red; nitrogen, dark blue; sulfur, yellow). Glycine residues are depicted as blue spheres, and carbonyl oxygens are shown as red spheres. Hydrogen bonds are depicted as dotted lines.