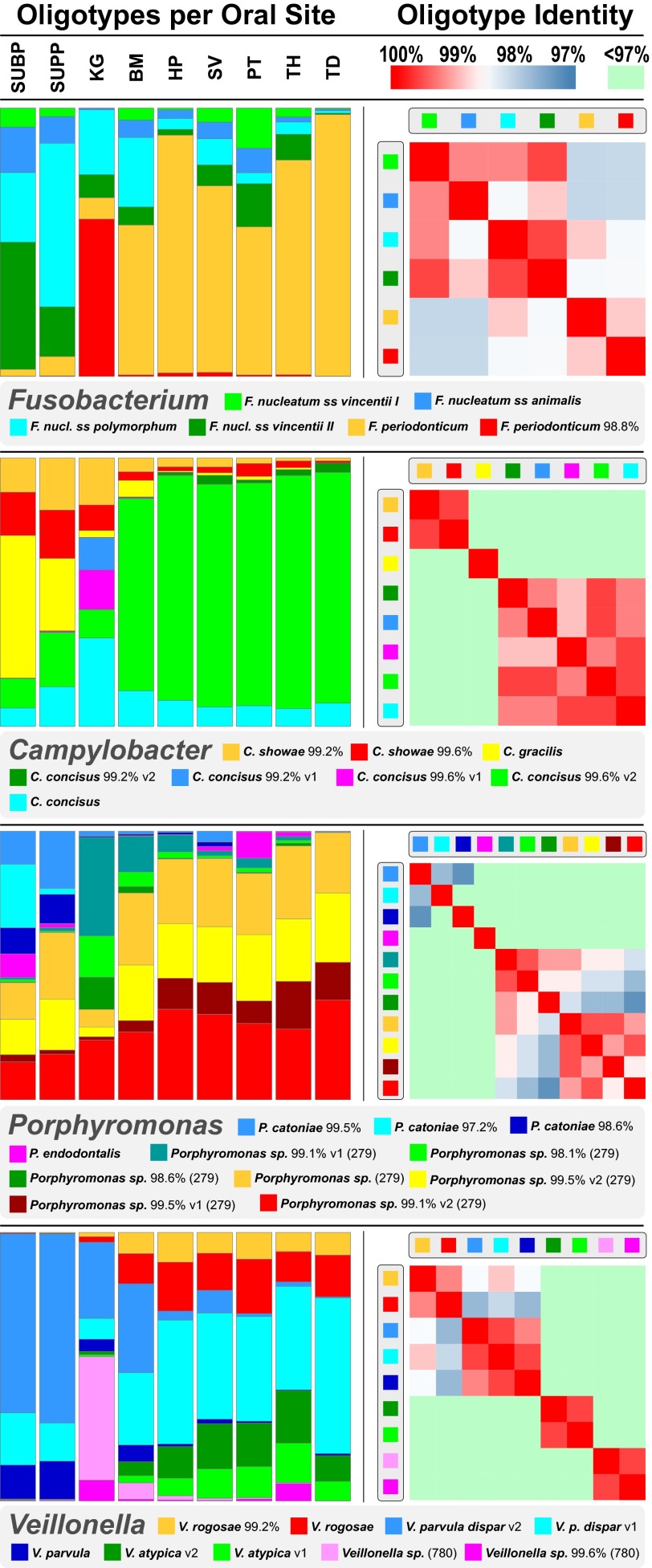
Fig. 3.
Abundant oligotypes of four oral genera showing habitat differentiation. Colored bars (Left) show the relative abundance of oligotypes of four oral genera in the V1-V3 data, averaged across individuals and shown for each sampling site. For simplicity, only oligotypes with a mean abundance of at least 0.5% (Fusobacterium, Veillonella), 0.2% (Porphyromonas), or 0.1% (Campylobacter) in at least one oral site are shown. Species names shown for oligotypes are the names of the identical sequence(s) in HOMD, or, for oligotypes not identical to any HOMD sequence, the names of the closest match sequence(s) followed by the percent identity to that match. Unnamed taxa with only a HOT designation are listed only when no named taxon is an exact or closest match; the HOT number is shown in parentheses. Heat maps (Right) show the percent nucleotide identity between each pair of oligotypes within a genus. Some oligotypes share the same name, followed by v1 or v2, because of the presence in HOMD of more than one reference sequence for these species.