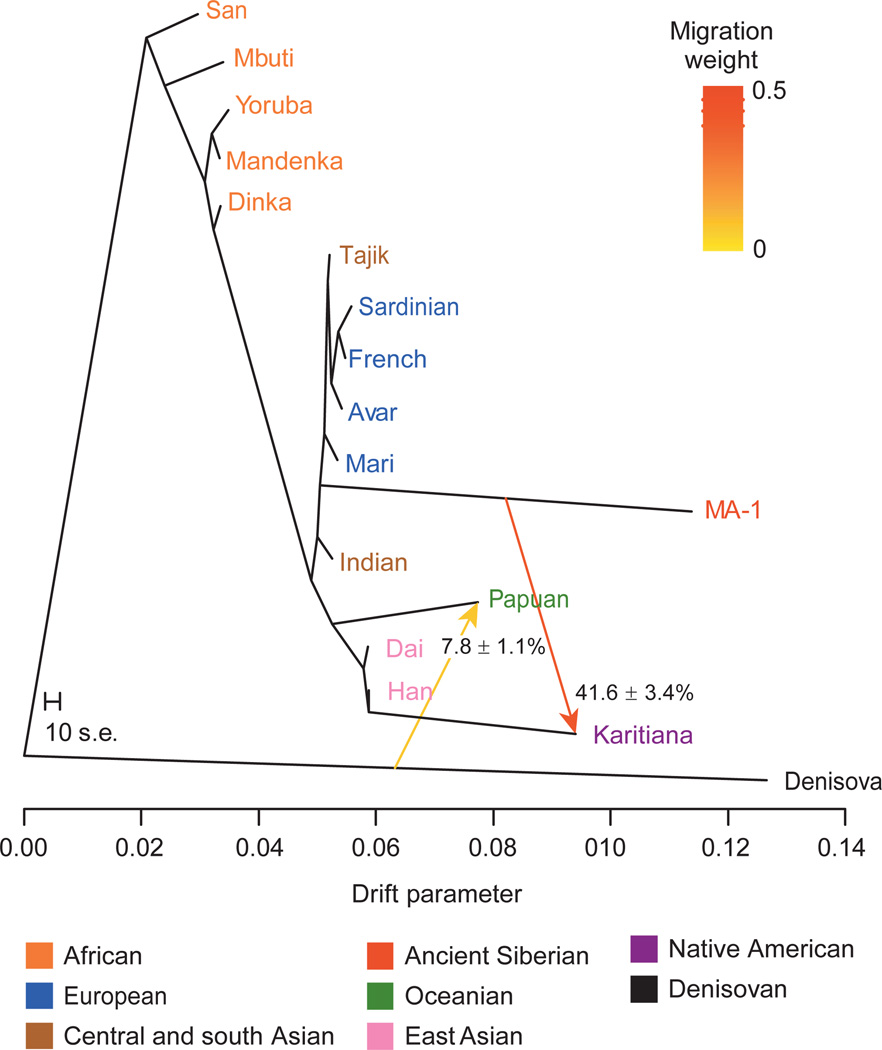
Figure 2.
Admixture graph for MA-1 and 16 complete genomes. An admixture graph with two migration edges (depicted by arrows) was fitted using TreeMix21 to relate MA-1 to 11 modern genomes from worldwide populations22, 4 modern genomes produced in this study (Avar, Mari, Indian and Tajik), and the Denisova genome22. Trees without migration, graphs with different number of migration edges, and residual matrices are shown in Supplementary Information, section 11. The drift parameter is proportional to 2Ne generations, where Ne is the effective population size. The migration weight represents the fraction of ancestry derived from the migration edge. The scale bar shows ten times the average standard error (s.e.) of the entries in the sample covariance matrix. Note that the length of the branch leading to MA-1 is affected by this ancient genome being represented by haploid genotypes.