Abstract
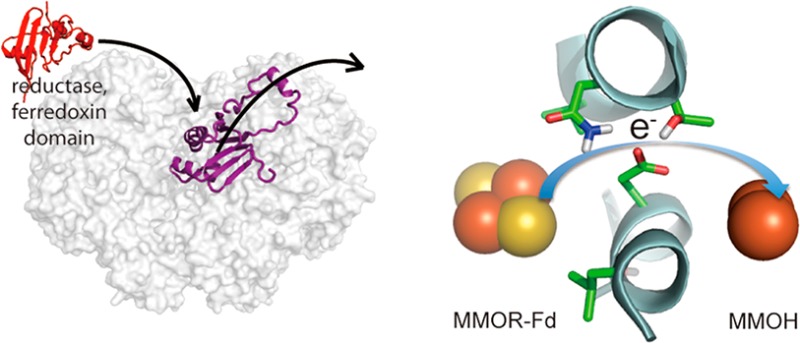
The hydroxylation or epoxidation of hydrocarbons by bacterial multicomponent monooxygenases (BMMs) requires the interplay of three or four protein components. How component protein interactions control catalysis, however, is not well understood. In particular, the binding sites of the reductase components on the surface of their cognate hydroxylases and the role(s) that the regulatory proteins play during intermolecular electron transfer leading to the hydroxylase reduction have been enigmatic. Here we determine the reductase binding site on the hydroxylase of a BMM enzyme, soluble methane monooxygenase (sMMO) from Methylococcus capsulatus (Bath). We present evidence that the ferredoxin domain of the reductase binds to the canyon region of the hydroxylase, previously determined to be the regulatory protein binding site as well. The latter thus inhibits reductase binding to the hydroxylase and, consequently, intermolecular electron transfer from the reductase to the hydroxylase diiron active site. The binding competition between the regulatory protein and the reductase may serve as a control mechanism for regulating electron transfer, and other BMM enzymes are likely to adopt the same mechanism.
Introduction
Bacterial multicomponent monooxygenases (BMMs) comprise a family of enzymes capable of hydroxylating or epoxidizing a wide range of hydrocarbons, including the greenhouse gas methane and environmentally hazardous substances such as benzene and trichloroethylene.1,2 BMM enzymes can be grouped into four classes: the three-component enzymes soluble methane monooxygenases (sMMOs), phenol hydroxylases (PHs), and alkene monooxygenases (AMOs), and the four-component enzymes alkene/arene monooxygenases.1 All BMM enzymes contain three common components: a hydroxylase, a reductase, and a regulatory protein. Alkene/arene monooxygenases require an additional Rieske protein for reducing the hydroxylase.1−4 The hydroxylase component is a multi-subunit dimeric (α2β2γ2 or α2β2) protein hosting a diiron center in each α-subunit. The carboxylate-bridged diiron center is the locus for O2 activation and subsequent substrate hydroxylation/epoxidation.3,4 It is similar to those in the R2 subunit of ribonucleotide reductase,5 ferritin,6 stearoyl acyl carrier protein Δ9 desaturase,7,8 and the aging-related protein Clk1.9,10 The reductase component is an NADH oxidoreductase with an [Fe2S2] cluster in the ferredoxin domain (Fd) and a flavin adenine dinucleotide (FAD) cofactor in the FAD domain, responsible for the reduction of the hydroxylase diiron center. The ultimate electron source is reduced nicotinamide adenine dinucleotide (NADH).11,12 The third component, a cofactor-less regulatory protein, couples NADH consumption to product formation.2,13,14 A key question regarding the catalytic mechanism of BMM enzymes is how component protein interactions achieve the timely control of electron transfer to the diiron active site, dioxygen activation, and hydrocarbon substrate oxidation.
Elucidating component protein binding sites on the hydroxylase is required as a foundation for answering such a question. An important clue comes from early crystallographic investigations of hydroxylases from the three-component system sMMO15,16 and the four-component toluene/o-xylene monooxygenase (ToMO).17 Both hydroxylases contain a structure of C2 symmetry with a shallow depression, termed the “canyon”, on each side of the protein dimer (Figure 1a). The canyon region was proposed as the docking site for the other component proteins,15,17 and, indeed, later crystallographic studies revealed that the regulatory component does occupy a portion of the canyon in hydroxylase–regulatory protein complexes of PH,18 toluene-4-monooxygenase (T4mO, another four-component BMM),19 and, very recently, sMMO (Figure 1b).20
Figure 1.
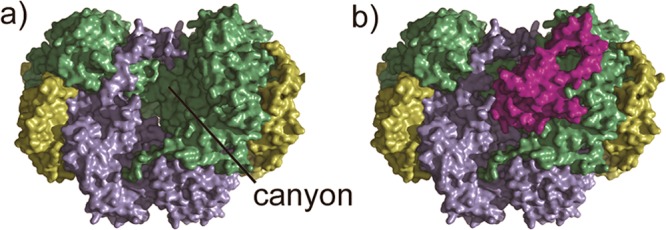
Crystal structure of the hydroxylase–regulatory protein complex of sMMO (PDB ID 4GAM): (a) the hydroxylase MMOH showing the canyon, and (b) MMOH in complex with the regulatory protein MMOB. There is another MMOB molecule binding to the canyon on the other side of MMOH. MMOH α-subunit is colored in green, β-subunit in blue, γ-subunit in yellow, and MMOB in purple.
The exact binding site of the reductase component has remained elusive, however. There is no crystal structure available for the hydroxylase–reductase complex of any BMM enzyme. By using the zero-length cross-linker 1-ethyl-3-(3-(dimethylamino)propyl)carbodiimide (EDC), a chemical cross-linking study of sMMO isolated from Methylosinus trichosporium OB3b revealed that the reductase, MMOR, cross-linked to the β-subunit of the hydroxylase MMOH, and that the regulatory component, MMOB, cross-linked to the α-subunit.21 A different result was obtained, however, using sMMO isolated from Methylococcus capsulatus (Bath), where either the full-length MMOR or its Fd cross-linked to the α-subunit using the same cross-linker, EDC.22 Further attempts to determine the binding site by identifying cross-linked residues failed. The two identified Fd cross-linking sites, Glu-56 and Glu-91, cross-linked to the N-terminal amino group of MMOH α-subunit, which is not observed in the crystal structure of MMOH owing to disorder.22
Because the MMOR binding site on MMOH is obscure, it was unclear how the regulatory protein and the reductase might interact in the complete enzyme system. Simulations of steady-state oxidase and oxygenase activities of sMMO as a function of component protein concentrations favored a non-competitive model, whereby MMOR and MMOB bind at distinct sites on MMOH,23 forming a hypothetical ternary complex. The formation of such a ternary complex was also proposed in a small-angle X-ray scattering (SAXS) study, where the species formed in the presence of large excess of MMOB and MMOR (10–20 equiv of each relative to MMOH) was modeled as a MMOH–2MMOB–2MMOR complex.24 Later crystallographic investigations of the hydroxylase–regulatory protein complexes, however, suggested that the regulatory component may block the reductase binding site,18−20 but there was no direct experimental evidence for such. The role of the regulatory protein in electron transfer from the reductase to the hydroxylase diiron center is also not well understood. A determination of the reductase binding site on the hydroxylase would clarify many of these questions.
Accordingly, in this study we determined the reductase binding site on the hydroxylase of sMMO isolated from Methylococcus capsulatus (Bath), by using hydrogen–deuterium exchange coupled to mass spectrometry (HDX-MS). The results clearly reveal that the Fd of MMOR indeed binds to the canyon of MMOH. More importantly, the Fd shares the same binding site as the core of MMOB; it therefore binds competitively with MMOB to MMOH. These conclusions are supported by computational docking and by binding competition assays. Consistent with the shared binding site, we show that MMOB does not facilitate, but actually inhibits, electron transfer. Overall, this work presents the first experimentally determined reductase Fd binding site on the hydroxylase of a BMM enzyme, and it reveals how the regulatory component may control electron transfer in the catalytic cycle.
Experimental Section
Materials
D2O was ordered from Cambridge Isotope Laboratories. 5-({2-[(Iodoacetyl)amino]ethyl}amino)naphthalene-1-sulfonic acid (IAEDANS) was obtained from Molecular Probes. Other chemicals were purchased from Sigma-Aldrich and used without further purification.
Protein Preparation
MMOH, MMOB, and MMOR were prepared as described previously.25 The expression system for MMOR ferredoxin domain (Fd, residues 1–107) was prepared by mutating S108 and F109 of wild-type MMOR to stop codons by site-directed mutagenesis, using primers shown in Table S1. Fd was expressed and purified as described previously,26 except that an additional step with a MonoQ column was employed to separate apo protein without the iron–sulfur cluster from the holo protein. The purified Fd had an A276nm/A330nm ratio of 1.05. MMOB D36C mutant was prepared as described previously.25 The expression system for the MMOB Δ2-33 D36C mutant was prepared by site-directed mutagenesis using MMOB D36C as the template; the primers are shown in Table S1. The MMOB Δ2-33 D36C protein was expressed and purified following the procedure described for wild-type MMOB.25
HDX-MS
HDX-MS was performed essentially as described.27 A 60 pmol portion of MMOH was incubated with Fd for a final MMOH:Fd concentration ratio of 1:6 during deuterium labeling. Under this condition, >95% of the Fd binding sites on MMOH were saturated, based on a Kd value of 0.9 μM. All mixtures were incubated for 20 min at room temperature before deuterium labeling. As a control, MMOH alone was incubated in 50 mM phosphate buffer (pH 7.0) and treated exactly the same as the Fd-bound protein. Deuterium exchange was initiated by dilution of each sample with 15-fold 50 mM phosphate buffer (pD 7.0), 99.9% D2O at room temperature. At each deuterium exchange time point (10 s, 1 min, 10 min, 60 min, 4 h, 6 h, and 8 h), an aliquot from the exchange reaction was removed and quenched by adjusting the pH to 2.5 with an equal volume of quench buffer (150 mM potassium phosphate buffer, H2O). Quenched samples were immediately frozen on dry ice and stored at −80 °C until analysis. Several undeuterated control samples were prepared in the same way as the deuterium-labeled samples and were used for validation of the peptic peptides of the proteins used in the deuterium labeling experiments.
Each flash-frozen sample was rapidly thawed and injected into a Waters nanoACQUITY with HDX Technology (Waters Corp.).28 The protein samples were digested online using a 2.1 mm × 30 mm Poroszyme immobilized pepsin cartridge (Applied Biosystems). The digestion temperature was set to 15 °C and the digestion was performed for 30 s. The cooling chamber of the ultra-performance liquid chromatography (UPLC) system, which housed all the chromatographic elements, was held at 0.0 ± 0.1 °C for the entire time of the measurements. The injected peptides were trapped and desalted for 3 min at 100 μL/min and then separated in 14 min by a 5% to 40% acetonitrile:water gradient at 40 μL/min. The separation column was a 1.0 × 100.0 mm ACQUITY UPLC C18 bridged ethyl hybrid particles (BEH) column (Waters Corp.) containing 1.7 μm particles, and the back pressure averaged 8800 psi at 0.1 °C. The average amount of back-exchange using this experimental setup was 18–25%, based on analysis of highly deuterated peptide standards. Deuterium levels were not corrected for back-exchange and are therefore reported as relative;29 however, all comparison experiments were done under identical experimental conditions, thus negating the need for a back exchange correction.29 The UPLC step was performed with protonated solvents, thereby allowing deuterium to be replaced with hydrogen from side chains and the amino/carboxyl terminus that exchange much more rapidly than amide linkages.30 All experiments were performed in triplicate. The average error in determining the deuterium levels was ±0.1 Da in this experimental setup, consistent with previously obtained values.31 In order to eliminate peptide carryover, a wash solution of 1.5 M guanidine hydrochloride, 0.8% formic acid, and 4% acetonitrile was injected after each run.
Mass spectra were obtained with a Waters XEVO G2 TOF instrument equipped with standard electrospray ionization source (Waters Corp.). The instrument configuration was the following: capillary was 3.2 kV, trap collision energy at 6 V, sampling cone at 35 V, source temperature of 80 °C, and desolvation temperature of 175 °C. Mass spectra were acquired over an m/z range of 100–1900. Mass accuracy was ensured by calibration with 500 fmol/μL human [Glu1]-Fibrinopeptide B and was less than 10 ppm throughout all experiments. The mass spectra were processed with the software DynamX 2.0 (Waters Corp.) by centroiding an isotopic distribution corresponding to the +2, +3, or +4 charge state of each peptide. Deuteration levels were calculated by subtracting the centroid of the isotopic distribution for peptide ions of undeuterated protein from the centroid of the isotopic distribution for peptide ions from the deuterium-labeled sample. The resulting relative deuterium levels were automatically plotted versus the exchange-in time. Identification of the peptic fragments was accomplished through a combination of exact mass analysis and MSE using Identity Software (Waters Corp.). MSE was performed by a series of low–high collision energies ramping from 5 to 32 V, therefore ensuring proper fragmentation of all the peptic peptides eluting from the LC system.32 Peptic maps were obtained with DynamX 2.0 software (Waters Corp.).
Fluorescent Labeling and Fluorescence Anisotropy Measurements
IAEDANS-labeled MMOB D36C and Δ2-33 D36C mutants were prepared following procedures described previously.25 Concentrations of the labeled proteins were determined by using the Bradford assay (Bio-Rad). The excitation wavelength was set to 336 nm and emission was monitored at 490 nm. Samples were made in 25 mM MOPS, pH 7.0 buffer; the concentration of fluorescently labeled protein was 1 μM.
Simulations of the Fluorescence Anisotropy Titration Curves
A competitive model was used to simulate titration curves. MMOH was considered to have two non-interacting binding sites (Hsite). The simulation procedures are described as follows, taking the titration of Fd into 1 μM MMOH and 1 μM IAEDANS-labeled MMOB as an example. Two equilibria were considered, eqs 1 and 2, where [Hsite], [B], and [Fd] are the concentrations of free MMOH binding site, free MMOB, and free Fd; [Hsite-B] and [Hsite-Fd] are the concentrations of bound MMOB and Fd; [Hsite]total, [B]total, and [Fd]total are the total concentrations of MMOH binding site (2 μM), MMOB (1 μM), and Fd (the total amount titrated in). The Kd for the H–B complex (Kd,H–B) was determined previously to be 0.55 μM;25 several Kd values for the H–Fd complex (Kd,H–Fd) were tested to allow us to choose the one that best simulated the experimental data.
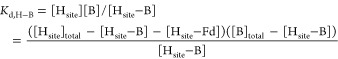 |
1 |
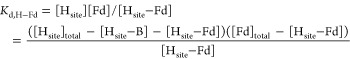 |
2 |
The concentrations [B] and [Hsite–B] were first calculated for each titration point by numerically solving the simultaneous eqs 1 and 2. These values were then used to calculate the observed fluorescence anisotropy robs (eq 3), which is the sum of fluorescence anisotropy of free and bound MMOB weighted by their fractional fluorescence intensity,33
| 3 |
where rB and rHsite–B are the fluorescence anisotropy of free and bound MMOB, and fB and fHsite–B are the fractional fluorescence intensity of free and bound MMOB, respectively. The fB and fHsite–B parameters can be expressed in terms of [B], [Hsite–B], and the molar fluorescence intensity of free and bound MMOB, FB and FHsite–B:
| 4 |
| 5 |
Substituting eqs 4 and 5 into eq 3, the fluorescence anisotropy of each titration point can be calculated on the basis of [B] and [Hsite–B] by solving eqs 1 and 2:
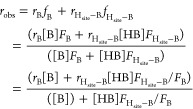 |
6 |
The same simulation procedures were followed for the titration of Fd into 1 μM MMOH and 1 μM IAEDANS-labeled MMOB Δ2-33. The Kd value of 2.67 μM for the H–B Δ2-33 complex determined in this study was used for the simulation (Figure S3). Several Kd values (0.9, 2, and 6 μM) for the H–Fd complex (Kd,H–Fd) were tested to see which one best fits the experimental data.
Chemical Cross-Linking
To a mixture of 10 μM MMOH, 20 μM MMOR, and 0–120 μM MMOB in 50 mM MOPS buffer, pH 7.0, was added 10 mM cross-linker EDC. The reaction was incubated at room temperature for 10 min and then quenched by adding an equal volume of SDS loading buffer.22 The reaction was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Electron Transfer Studies
The electron transfer kinetics of sMMO were studied by stopped-flow optical spectroscopy at 15 °C. The tubing and syringes of the Hi-Tech Scientific SF-61 DX2 double-mixing stopped-flow instrument were made anaerobic by first flushing and then incubating with 15 mM anaerobic sodium dithionite solution for 3 h, followed by flushing with 25 mM anaerobic MOPS buffer, pH 7.0 right before use. The following steps were performed inside of a glovebox with an O2 level of less than 0.5 ppm. To investigate electron transfer from chemically reduced reductase, 40 μM degassed Fd or MMOR was titrated with 3 mM sodium dithionite until the absorption at 405 nm no longer changed, and the resulting sample was then loaded into a gastight syringe. A 20 μM quantity of MMOH, or 20 μM MMOH premixed with 40 μM MMOB in the presence or absence of 10 μM MMOR, was degassed and sealed in another gastight syringe. To study electron transfer from NADH, a 40 μM solution of anaerobic NADH was sealed in one syringe; 20 μM MMOH premixed with 40 μM MMOR in the presence/absence of 40 μM MMOB was sealed in another syringe. The syringes were then taken out from the glovebox and connected to the stopped-flow instrument. Equal volumes of reagents from each syringe were rapidly mixed by the stopped-flow instrument, and the electron transfer kinetics were monitored by recording the absorbance change at 470 or 458 nm. Data were fit by two (when chemically reduced Fd or MMOR was used as the electron source) or three (when NADH was used as the electron source) exponentials, and effective electron transfer rates were calculated as weighted averages of individual electron transfer rate constant.
Results and Discussion
HDX-MS Study of MMOH
HDX-MS is a powerful tool for probing protein structure, dynamics, and the binding interface.34,35 The rationale behind HDX-MS relies on protein backbone amide protons that are in constant exchange with solvent protons, or deuteriums if in deuterated solvent. The number of exchangeable protons and their rates of exchange depend on factors such as pH, temperature, chemical environment, and the three-dimensional protein architecture,30,34−37 thus reflecting the structure and dynamics of the protein. Typically, protein backbone amide protons exchange rapidly with deuterons if they are involved in weak or suboptimal hydrogen bonds, reside at/near the surface, or are readily accessible to the solvent; the exchange rates are slower if they are involved in strong intramolecular hydrogen bonds and/or are less accessible to solvent.38 HDX-MS has also been successfully applied to determine protein–protein binding sites, based on the reduced solvent exposure in regions that constitute the binding interface.35,36,40
Here we first probed the dynamics of MMOH alone by HDX-MS. MMOH is a 251.3 kDa homodimer that consists of three protomer subunits in each monomer: α (60.6 kDa), β (45.1 kDa), and γ (19.8 kDa). Upon pepsin digestion, 165 overlapping MMOH peptic peptides were detected, covering 93.9%, 93.5%, and 96.4% of the sequences of the α-, β-, and γ-subunits, respectively (Figure S1). The deuterium incorporation and protein dynamics were followed from 10 s up to 8 h. All the peptic peptides that were followed by HDX-MS are displayed in Figure S2. Most of these peptides showed low deuterium uptake even after an 8 h incubation in deuterated buffer (Figure 2). The data indicate very slow dynamics in most of the three subunits of the homodimer, suggesting that the protein is very rigid and not undergoing breathing movements indicative of a dynamic and solvent exposed structure.29,35 Amide backbone hydrogens involved in hydrogen-bonding interactions in secondary structural elements such as α-helices and β-sheets exhibit slow exchange rates.41 Therefore, the low deuterium uptake is consistent with the high helical content of MMOH.15,16,39 The α and β-subunits are more protected from exchange compared with the γ-subunit, the peptides of which seem to indicate a more accessible and dynamic structure (Figure 2).
Figure 2.
Summary of HDX-MS data for free MMOH in solution at four time points. The HDX-MS data are mapped onto PDB entry 1MTY,39 with the color code indicated for deuteration times shown at the bottom of each image. The HDX-MS data are shown only on one monomer; the second monomer is represented in sand color.
HDX-MS Study of the MMOH–Fd Complex
We next used HDX-MS to locate regions of MMOH that show differences in exchange upon binding to the ferredoxin domain (Fd, residues 1–107, 11.8 kDa) of MMOR. MMOR is characterized by a modular structure containing both FAD and ferredoxin domains.42 The two isolated domain proteins both have stable structures43,44 and retain the biochemical properties of the two domains in full-length MMOR.42 Electron transfer to the diiron(III) centers in the hydroxylase involves first, NADH reductions of the oxidized FAD cofactor to its hydroquinone form, followed by the [Fe2S2] cluster in the Fd sequentially shuttling two electrons from the reduced FAD cofactor to the diiron center in MMOH.12,23,45 The Fd protein is a smaller yet competent model of MMOR for studying electron transfer as well as binding to MMOH.26
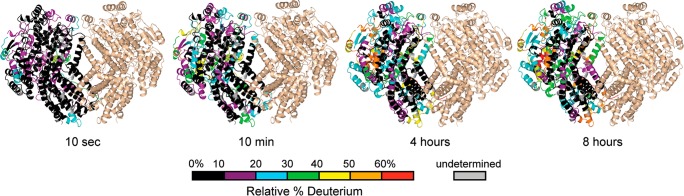
Among the 165 detected peptic peptides of MMOH in the presence of Fd (Figure S2), six peptides derived from the α-subunit and three from the β-subunit exhibited lower deuterium uptake compared with samples without Fd (Figures 3 and S2), covering residues 70–88 and 236–255 of the α-subunit, and residues 37–48 of the β-subunit. The differences in deuterium uptake were visible after ∼10–60 min deuteration but not at earlier time points, possibly because the exchange rates in the absence of Fd were already very slow due to stable H-bonds in the α-helices. In the presence of Fd the reduced solvent exposure at the binding site modestly reduced the exchange rates. The differences in deuterium uptake in the presence and absence of Fd were subtle even at longer incubation times (up to 8 h exchange), but were consistent in the set of overlapping peptides. Typically, differences in deuteration greater than 0.4 Da but less than 1 Da are considered subtle. The experiments were performed in triplicate, and the average error of the measurements was ±0.1 Da. All other MMOH peptic peptides exhibited the same deuterium uptake in the presence or absence of Fd (Figure S2).
Figure 3.
Hydrogen–deuterium exchange kinetics for four representative peptides that showed different deuterium uptake in the presence (blue traces) or absence (red traces) of Fd: (a) residues 70–81, α-subunit; (b) residues 82–88, α-subunit; (c) residues 237–242, α-subunit; and (d) residues 40–47, β-subunit. The largest number on the y-axis represents the maximum amount of deuterium that can be incorporated in each peptic peptide.
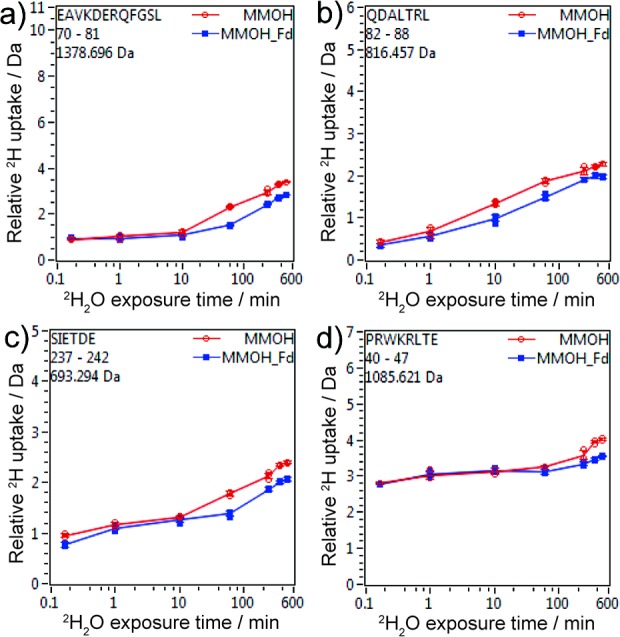
We then mapped the peptides that showed decreased deuterium uptake in the presence of Fd onto the crystal structure of MMOH (PDB entry 4GAM). These peptides cluster in the canyon region at the α2β2 interface, representing a possible Fd binding site (Figure 4a). This region includes the area closest to the diiron center from the protein surface, a preferred binding site for the iron–sulfur cluster of the reductase in order to facilitate fast electron transfer. This binding site determined by HDX-MS rationalizes the seemingly conflicting results of previous chemical cross-linking studies. Because the binding site consists of residues from both α- and β-subunits, MMOR can cross-link to either the α-subunit (for sMMO isolated from Methylococcus capsulatus (Bath))22 or the β-subunit (for sMMO isolated from Methylosinus trichosporium OB3b).21 It is unlikely that sMMOs isolated from different species have different reductase binding sites. The different cross-linking results are most likely due to different distributions of carboxyl groups and amino groups that are required to be in close proximity for the cross-linking mediated by EDC.
Figure 4.
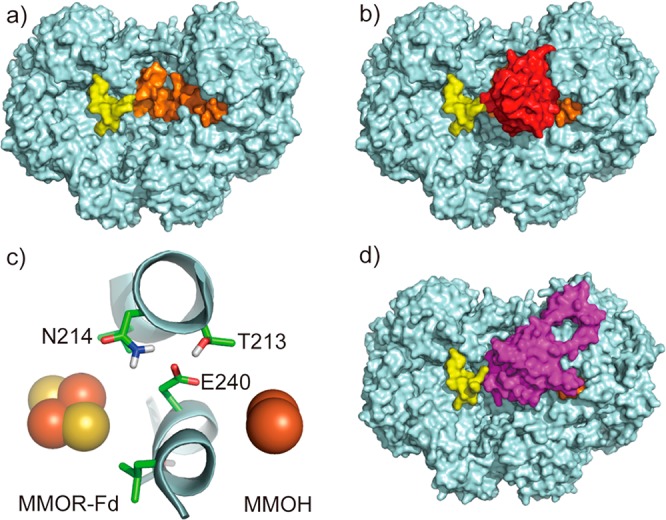
Fd and MMOB binding sites on MMOH. (a) MMOH peptides that showed decreased deuterium uptake in the presence of Fd are mapped onto the crystal structure of MMOH (PDB ID 4GAM). (b) A computationally docked MMOH–Fd complex. (c) A closer view of the binding interface in the docked model: the [Fe2S2] cluster of Fd, MMOH residues at the binding interface, and the diiron center of MMOH, viewed from the top of the figure. (d) The crystal structure of MMOH-2MMOB, showing that MMOB covers the Fd binding site. MMOH is colored in cyan; binding site peptides of the α-subunit in orange and those of the β-subunit in yellow; Fd in red; and MMOB in purple.
To validate the HDX-MS-determined Fd binding site, we performed computational docking using the ClusPro server.46−49 The structure of Fd (residues 1–98, PDB ID 1J4Q) previously determined by solution-state NMR spectroscopy43 and the crystal structure of MMOH (PDB ID 4GAM)39 were used as input structures, and the NMR-determined Fd residues involved in binding to MMOH43 were used as docking constraints. The results show that Fd covers the HDX-MS-determined binding surface (Figure 4b). The β-subunit peptides involved in binding (yellow region in Figure 4) seem uncovered in the docked structure (Figure 4b), but upon a closer examination residues 45–48 inside the canyon were found to be in close contact with Fd. The docking result is therefore in full agreement with the HDX-MS data.
Implications for Electron Transfer from MMOR to MMOH
A close examination of the docked MMOH–Fd complex reveals that the diiron active site of MMOH is approximately 14 Å away from the [Fe2S2] cluster of Fd, a favorable distance for electron transfer.24 Consistent with HDX-MS results, the docked model reveals that Fd covers the pore region of MMOH, which we previously proposed as the proton transfer pathway from the MMOH surface to its diiron center.20,50 Residue E240 in the MMOH α-subunit, the gating residue of the pore, is situated midway between the [Fe2S2] cluster of Fd and the diiron center of MMOH (Figure 4c). Previously, this residue was found to shift its conformation toward the protein interior upon MMOB binding, closing the pore and possibly bringing in a proton to the diiron active site.20 We propose that an identical conformational change occurs when MMOR binds to MMOH during electron transfer from the Fd [Fe2S2] cluster to the carboxylate-bridged diiron center, providing the mechanism for proton-coupled electron transfer.
Binding Competition between the Reductase and the Regulatory Protein
Strikingly, the Fd binding site determined here overlaps largely with that of MMOB as previously determined by the crystal structure of the MMOH–2MMOB complex. In particular, this structure reveals that the core of MMOB docks into the canyon region, covering the very same area where we now conclude Fd binds, while the N-terminal tail of MMOB binds to an adjacent location on MMOH, on the surface of helices H and 4 of the α-subunit, adopting a ring-like conformation (Figure 4d).20 This observation requires the Fd of MMOR and the core of MMOB compete for the same binding site in the canyon of MMOH.
To test this possibility, we first investigated the binding competition between Fd and the core of MMOB by fluorescence anisotropy titrations. The MMOB core was prepared by truncating the N-terminal tail at residue 33 (Δ2-33), and a cysteine mutation D36C was introduced in order to attach the fluorophore IAEDANS. The N-terminal truncated MMOB (designated MMOB Δ2-33) is still able to bind to MMOH but with lower affinity (Kd = 2.67 μM, Figure S3; Kd = 0.55 μM for the full-length MMOB25). To characterize the binding competition, Fd was titrated into a mixture of 1 μM MMOH and 1 μM IADEANS-labeled MMOB Δ2-33. The fluorescence anisotropy decreased steadily as increasing amounts of Fd were added (Figure 5a), indicating displacement of N-terminal truncated MMOB by Fd from MMOH (Scheme 1a). The titration curve could be simulated by assuming that the N-terminal truncated MMOB and Fd compete for the same binding sites on MMOH (MMOHsite, one on each side of MMOH), and an apparent Kd value of 2 μM was obtained for the MMOH–Fd complex on the basis of the simulations (Figure 5a).
Figure 5.
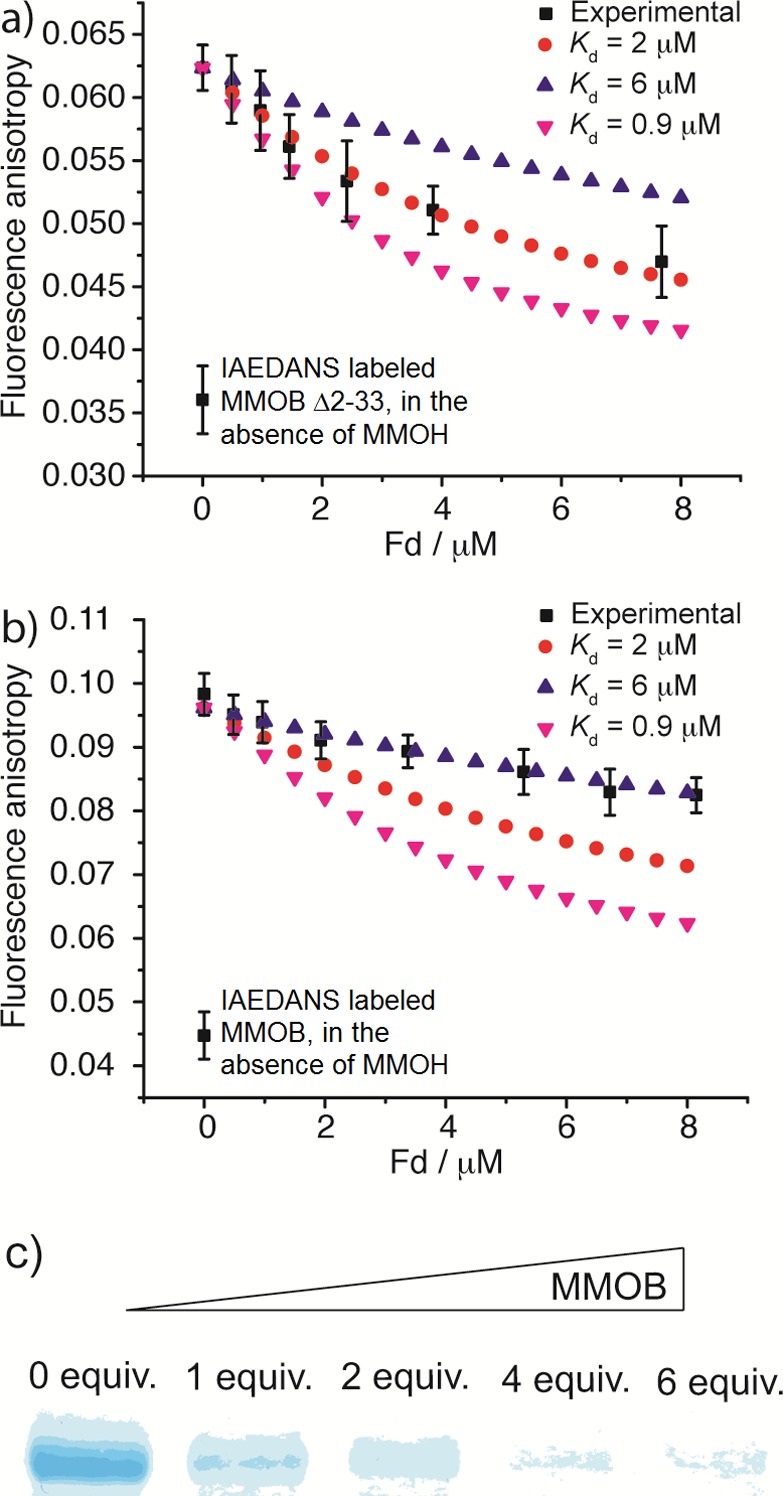
Experimental evidence for MMOR and MMOB binding competition. (a,b) Titrating Fd into 1 μM MMOH and 1 μM IADEANS-labeled N-terminal truncated (Δ2–33) MMOB (a) or full-length MMOB (b). The titration curves are simulated assuming Kd = 0.9, 2, and 6 μM for the MMOH–Fd complex; Kd = 2 and 6 μM best fit the experimental data in (a) and (b), respectively. (c) MMOB inhibits MMOR cross-link to MMOH. EDC was used as the cross-linking reagent, and 0, 1, 2, 4, and 6 equiv MMOB relative to MMOR were added to a mixture of 10 μM MMOH and 20 μM MMOR. The cross-linking reaction mixtures were resolved by SDS-PAGE, and bands corresponding to the MMOR−α-subunit cross-linking product are shown.
Scheme 1. Schematic Representations for MMOR Ferredoxin Domain (Red) and MMOB (Purple) Binding Competition for the Canyon of MMOH (Gray): (a) Binding Competition between Fd and MMOB Core (MMOB Δ2-33) and (b) Binding Competition between Fd and Full-Length MMOB.
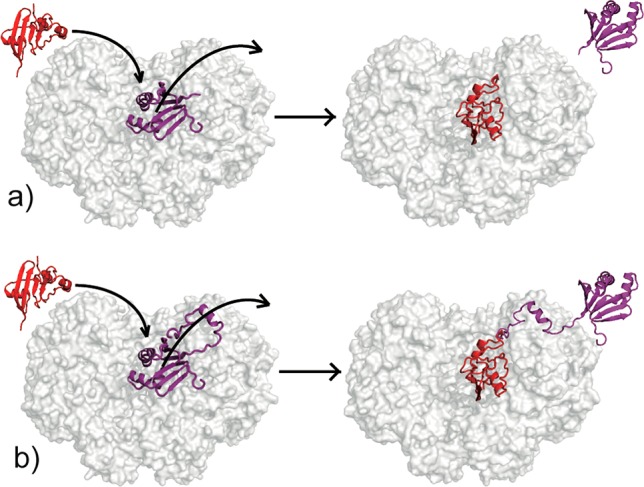
In the binding competition between Fd and full-length MMOB (b), Fd may displace the core of MMOB from the canyon, but the N-terminal tail may still bind to MMOH.
The apparent Kd value increased to 6 μM for the Fd–MMOH complex when the titration experiment was repeated with the full-length MMOB labeled with IAEDANS (Figure 5b), indicating that Fd is less effective in displacing full-length MMOB. This result is an expected consequence of the additional binding site of the N-terminal tail of MMOB on the surface of helices H and 4 of the MMOH α-subunit,20 a binding region not shared with Fd. It is possible that the N-terminal tail serves as an anchor attached to the surface of MMOH, such that, when Fd displaces the core of MMOB from the canyon, MMOB does not completely dissociate from MMOH (Scheme 1b).20 A simple competitive binding model would therefore no longer apply to the component interactions in this case. Similar results were obtained by using the full-length MMOR as the titrant. An apparent Kd value of 8 μM for the MMOH–MMOR complex is required to fit the titration curve using a competitive binding model (Figure S4), considerably larger than the 0.9 μM Kd value previously determined23 for the MMOH–MMOR complex in the absence of MMOB.
These results clearly demonstrate that both Fd and the full-length MMOR are able to displace MMOB, in particular the core of MMOB, from MMOH. Conversely, MMOB can displace MMOR from MMOH. This property was demonstrated by studying the effect of MMOB on MMOR cross-linking to MMOH. As shown in Figure 5c, MMOR forms cross-links to the MMOH α-subunit in the presence of EDC, as demonstrated previously;22 the yield of this cross-link decreases in a dose-dependent manner as the MMOB concentration increases from 0 to 6 equiv relative to the MMOR concentration, indicating that MMOB blocks MMOR binding, and thus its cross-linking, to MMOH. A similar finding was reported for sMMO from M. trichosporium OB3b.21
MMOB Inhibits Electron Transfer
The determination of component protein binding sites on the hydroxylase and their binding competition form a basis for understanding the role of the regulatory protein in electron transfer reactions of sMMO. Previously, MMOB was proposed to facilitate electron transfer from MMOR to MMOH, and this effect was ascribed to formation of a hypothetical MMOH–2MMOB–2MMOR ternary complex.23 It was hypothesized that the increased electron transfer exhibited by this ternary complex is due to a conformational change of MMOH induced by MMOR. Considering the substoichiometric concentration of MMOR found in vivo and used during activity assays,23,51 it was further proposed that such conformational changes are retained throughout the catalytic cycle even after MMOR dissociates from the ternary complex.26 Considering our current finding that the core of MMOB and the Fd of MMOR compete for binding to the canyon of MMOH, these proposals are deemed unlikely. Because MMOB can inhibit MMOR binding to the canyon, MMOB would inhibit electron transfer as well.
To test experimentally whether MMOB increases the electron transfer rate, and whether pre-equilibrating MMOR with other protein components would result in such an increase, we designed two sets of single-turnover, single-mixing stopped-flow experiments. In the first set, the electron transfer reactions were initiated by mixing 2 equiv of chemically reduced Fd to (i) 1 equiv of oxidized MMOH alone, (ii) 1 equiv of oxidized MMOH equilibrated with 2 equiv of MMOB, and (iii) 1 equiv of oxidized MMOH equilibrated with 2 equiv of MMOB as well as 0.5 equiv of MMOR (Figure 6a). The reactions were monitored at 470 nm, which increases in intensity as electrons are transferred to the MMOH diiron sites from the reduced [Fe2S2] clusters. The previous proposal would predict that case (iii), where all three components were pre-mixed and equilibrated, would exhibit the fastest electron transfer rate. The actual results (Figure 6b), however, showed that reaction (i) displays the fastest electron transfer rate. Including MMOB in reactions (ii) and (iii) significantly retarded the electron transfer reaction, even when MMOR was added and pre-equilibrated with other components. The effective electron transfer rates under different conditions are summarized in Table 1. These experiments were repeated using chemically reduced full-length MMOR as the reductant, and the same trend was observed (Figure S5).
Figure 6.
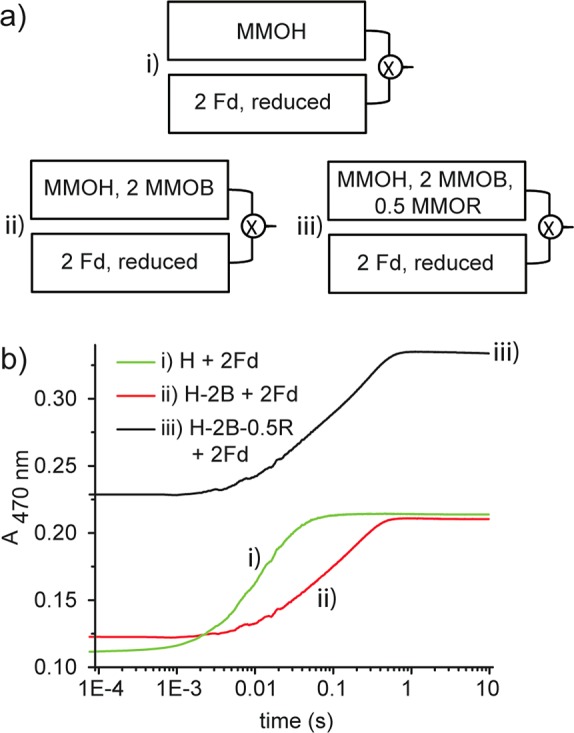
MMOB inhibits electron transfer, using chemically reduced Fd as the electron source. (a) Schematic diagrams for the first set of experiments. (b) Electron transfer kinetic curves. T = 15 °C.
Table 1. Effective Electron Transfer Ratesa.
| syringe 1 | syringe 2 | effective electron transfer rate (s–1) |
|---|---|---|
| 2Fd, reduced | MMOH | 76.1 ± 1.3 |
| 2Fd, reduced | MMOH–2MMOB | 17.0 ± 0.7 |
| 2Fd, reduced | MMOH–2MMOB–0.5MMOR | 18.3 ± 0.8 |
| 2NADH | MMOH–2MMOR | 14.0 ± 0.4 |
| 2NADH | MMOH–2MMOB–2MMOR | 3.52 ± 0.31 |
In the second set of experiments, the electron transfer reaction was initiated by mixing 2 equiv of NADH with (i) 1 equiv of oxidized MMOH and 2 equiv of MMOR and (ii) 1 equiv of oxidized MMOH, 2 equiv of MMOB, and 2 equiv of MMOR (Figure 7a). The reactions were monitored at 470 nm, where the absorption first decreased due to the reduction of the FAD cofactor by NADH and the subsequent intra-MMOR electron transfer to the [Fe2S2] cluster.12 The absorption at this wavelength then increased, as electrons were transferred from these cofactors to the diiron centers in MMOH. The previous proposal would predict that reaction (ii), which had all three protein components pre-equilibrated, would display the faster electron transfer rate, but in fact it is slower (Figure 7b), again demonstrating the inhibitory effect of MMOB on electron transfer (Table 1).
Figure 7.
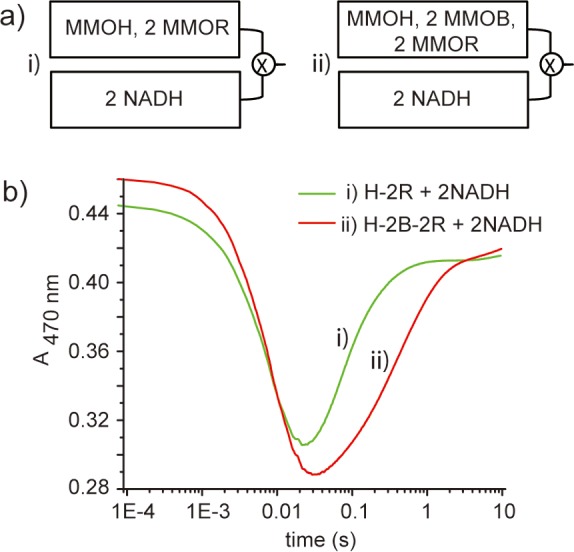
MMOB inhibits electron transfer, using NADH as the electron source. (a) Schematic diagrams for the second set of experiments. (b) Electron transfer kinetic curves.
Binding Competition: A Possible Mechanism for Modulating Electron Transfer
The inhibitory effect of MMOB on electron transfer is in full agreement with our finding that the core of MMOB shares a binding site on MMOH with the Fd of MMOR such that MMOB inhibits MMOR binding. Although electron transfer is retarded, it is still much faster than the rate of substrate turnover (kcat = 0.1 s–1 at 15 °C using propylene as substrate). The advantage of such competitive binding is that it provides a mechanism for modulating electron transfer during catalysis. By fine-tuning the affinity of MMOR and MMOB for the canyon region of MMOH, the binding of MMOR can be inhibited during dioxygen activation, preventing undesired electron transfer that could quench activated dioxygen intermediate species. Such quenching is suggested by oxidase activity displayed by MMOH and MMOR in the absence of MMOB.23
The modulation of electron transfer exhibited by the MMOB regulatory protein is most likely a feature common to other BMM enzymes. These enzymes share conserved protein sequences as well as structures,1,2 and, like sMMO, their regulatory proteins also bind to an analogous canyon region of the hydroxylase, as illustrated in the X-ray structures of the protein complexes in PH18 and T4mO,19 blocking the shortest pathway to the hydroxylase diiron center from its surface. The reductase may therefore need to compete with the regulatory protein for binding in order to deliver electrons to the diiron centers in these enzymes as well. In disagreement with this hypothesis, previous studies of PH and ToMO reported accelerated electron transfer in the presence of the regulatory protein.52 These electron transfer rates, however, were determined in steady-state assays in the absence of substrates, by measuring NADH consumption. Under these conditions, the NADH consumption rate depends on the oxidase activity of the hydroxylase, and the accelerated NADH consumption in the presence of the regulatory component is most likely due to increased oxidase activity and not an indication of accelerated electron transfer.
Conclusions
Using HDX-MS, the binding site for the ferredoxin domain of MMOR was determined to be in the canyon of MMOH, the same region where the core of the MMOB regulatory protein binds. This finding is consistent with previous chemical cross-linking results and the current computational docking study, as well as a series of binding competition assays. MMOB inhibits MMOR binding to the canyon as well as the electron transfer that leads to reduction of the hydroxylase. The previous proposals that MMOB increases the electron transfer rate when all three components are pre-equilibrated has been tested experimentally and proved to be invalid. Regulatory proteins of other BMM enzymes may similarly share binding sites with their reductases and inhibit electron transfer.
Such binding competition would provide a control mechanism for electron transfer in BMM enzymes. To initiate the catalytic cycle, the reductase displaces the regulatory protein from the canyon of the hydroxylase, reducing the diiron center from diiron(III) to diiron(II); the regulatory protein then re-binds to the canyon and displaces the reductase, initiating O2 activation and substrate oxidation. The binding affinities of the component proteins may be fine-tuned so that the reductase is unable to displace the regulatory protein at this step, preventing quenching of activated oxygen intermediates P*, Hperoxo, and Q.53 At the end of the catalytic cycle, the diiron center returns to the diiron(III) state, and the reductase can bind again to the canyon, priming the enzyme for the next cycle of catalysis.
Acknowledgments
We thank Ali D. Liang for assistance with stopped-flow experiments and for helpful discussions. This work was supported by NIH Grants GM032134 for S.J.L., and GM086507 and GM101135 for J.R.E., and a research collaboration with the Waters Corporation.
Supporting Information Available
Figures S1–S5 and Table S1. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Leahy J. G.; Batchelor P. J.; Morcomb S. M. FEMS Microbiol. Rev. 2003, 27, 449. [DOI] [PubMed] [Google Scholar]
- Notomista E.; Lahm A.; Di Donato A.; Tramontano A. J. Mol. Evol. 2003, 56, 435. [DOI] [PubMed] [Google Scholar]
- Sazinsky M. H.; Lippard S. J. Acc. Chem. Res. 2006, 39, 558. [DOI] [PubMed] [Google Scholar]
- Murray L. J.; Lippard S. J. Acc. Chem. Res. 2007, 40, 466. [DOI] [PubMed] [Google Scholar]
- Nordlund P.; Dalton H.; Eklund H. FEBS Lett. 1992, 307, 257. [DOI] [PubMed] [Google Scholar]
- Hempstead P. D.; Hudson A. J.; Artymiuk P. J.; Andrews S. C.; Banfield M. J.; Guest J. R.; Harrison P. M. FEBS Lett. 1994, 350, 258. [DOI] [PubMed] [Google Scholar]
- Fox B. G.; Shanklin J.; Ai J.; Loehr T. M.; Sanders-Loehr J. Biochemistry 1994, 33, 12776. [DOI] [PubMed] [Google Scholar]
- Kurtz D. M. Jr. J. Biol. Inorg. Chem. 1997, 2, 159. [Google Scholar]
- Behan R. K.; Lippard S. J. Biochemistry 2010, 49, 9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T.-T.; Lee S. J.; Apfel U.-P.; Lippard S. J. Biochemistry 2013, 52, 2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund J.; Dalton H. Eur. J. Biochem. 1985, 147, 291. [DOI] [PubMed] [Google Scholar]
- Kopp D. A.; Gassner G. T.; Blazyk J. L.; Lippard S. J. Biochemistry 2001, 40, 14932. [DOI] [PubMed] [Google Scholar]
- Green J.; Dalton H. J. Biol. Chem. 1985, 260, 15795. [PubMed] [Google Scholar]
- Mitchell K. H.; Studts J. M.; Fox B. G. Biochemistry 2002, 41, 3176. [DOI] [PubMed] [Google Scholar]
- Rosenzweig A. C.; Frederick C. A.; Lippard S. J.; Nordlund P. Nature 1993, 366, 537. [DOI] [PubMed] [Google Scholar]
- Elango N. A.; Radhakrishnan R.; Froland W. A.; Wallar B. J.; Earhart C. A.; Lipscomb J. D.; Ohlendorf D. H. Protein Sci. 1997, 6, 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazinsky M. H.; Bard J.; Di Donato A.; Lippard S. J. J. Biol. Chem. 2004, 279, 30600. [DOI] [PubMed] [Google Scholar]
- Sazinsky M. H.; Dunten P. W.; McCormick M. S.; DiDonato A.; Lippard S. J. Biochemistry 2006, 45, 15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey L. J.; McCoy J. G.; Phillips G. N.; Fox B. G. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 19194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J.; McCormick M. S.; Lippard S. J.; Cho U. S. Nature 2013, 494, 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox B. G.; Liu Y.; Dege J. E.; Lipscomb J. D. J. Biol. Chem. 1991, 266, 540. [PubMed] [Google Scholar]
- Kopp D. A.; Berg E. A.; Costello C. E.; Lippard S. J. J. Biol. Chem. 2003, 278, 20939. [DOI] [PubMed] [Google Scholar]
- Gassner G. T.; Lippard S. J. Biochemistry 1999, 38, 12768. [DOI] [PubMed] [Google Scholar]
- Page C. C.; Moser C. C.; Chen X.; Dutton P. L. Nature 1999, 402, 47. [DOI] [PubMed] [Google Scholar]
- Wang W.; Lippard S. J. J. Am. Chem. Soc. 2014, 136, 2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazyk J. L.; Gassner G. T.; Lippard S. J. J. Am. Chem. Soc. 2005, 127, 17364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacob R. E.; Pene-Dumitrescu T.; Zhang J.; Gray N. S.; Smithgall T. E.; Engen J. R. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales T. E.; Fadgen K. E.; Gerhardt G. C.; Engen J. R. Anal. Chem. 2008, 80, 6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wales T. E.; Engen J. R. Mass Spectrom. Rev. 2006, 25, 158. [DOI] [PubMed] [Google Scholar]
- Englander S. W.; Kallenbach N. R. Q. Rev. Biophys. 1983, 16, 521. [DOI] [PubMed] [Google Scholar]
- Burkitt W.; O’Connor G. Rapid Commun. Mass Spectrom. 2008, 22, 3893. [DOI] [PubMed] [Google Scholar]
- Plumb R. S.; Johnson K. A.; Rainville P.; Smith B. W.; Wilson I. D.; Castro-Perez J. M.; Nicholson J. K. Rapid Commun. Mass Spectrom. 2006, 20, 1989. [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R.Principles of fluorescence spectroscopy, 3rd ed.; Springer: New York, 2006. [Google Scholar]
- Hoofnagle A. N.; Resing K. A.; Ahn N. G. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 1. [DOI] [PubMed] [Google Scholar]
- Konermann L.; Pan J.; Liu Y.-H. Chem. Soc. Rev. 2011, 40, 1224. [DOI] [PubMed] [Google Scholar]
- Woodward C. K.; Hilton B. D. Annu. Rev. Biophys. Bioeng. 1979, 8, 99. [DOI] [PubMed] [Google Scholar]
- Matthew J. B.; Richards F. M. J. Biol. Chem. 1983, 258, 3039. [PubMed] [Google Scholar]
- Resing K. A.; Hoofnagle A. N.; Ahn N. G. J. Am. Soc. Mass Spectrom. 1999, 10, 685. [DOI] [PubMed] [Google Scholar]
- Mandell J. G.; Falick A. M.; Komives E. A. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig A. C.; Brandstetter H.; Whittington D. A.; Nordlund P.; Lippard S. J.; Frederick C. A. Proteins: Struct., Funct. Bioinf. 1997, 29, 141. [PubMed] [Google Scholar]
- Printz M. P.; Williams H. P.; Craig L. C. Proc. Natl. Acad. Sci. U.S.A. 1972, 69, 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazyk J. L.; Lippard S. J. Biochemistry 2002, 41, 15780. [DOI] [PubMed] [Google Scholar]
- Müller J.; Lugovskoy A. A.; Wagner G.; Lippard S. J. Biochemistry 2002, 41, 42. [DOI] [PubMed] [Google Scholar]
- Chatwood L. L.; Müller J.; Gross J. D.; Wagner G.; Lippard S. J. Biochemistry 2004, 43, 11983. [DOI] [PubMed] [Google Scholar]
- Lund J.; Woodland M. P.; Dalton H. Eur. J. Biochem. 1985, 147, 297. [DOI] [PubMed] [Google Scholar]
- McCormick M. S.; Lippard S. J. Biochemistry 2011, 50, 11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau S. R.; Gatchell D. W.; Vajda S.; Camacho C. J. Nucleic Acids Res. 2004, 32, W96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau S. R.; Gatchell D. W.; Vajda S.; Camacho C. J. Bioinformatics 2004, 20, 45. [DOI] [PubMed] [Google Scholar]
- Kozakov D.; Brenke R.; Comeau S. R.; Vajda S. Proteins: Struct., Funct. Bioinf. 2006, 65, 392. [DOI] [PubMed] [Google Scholar]
- Kozakov D.; Beglov D.; Bohnuud T.; Mottarella S. E.; Xia B.; Hall D. R.; Vajda S. Proteins 2013, 81, 2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox B. G.; Froland W. A.; Dege J. E.; Lipscomb J. D. J. Biol. Chem. 1989, 264, 10023. [PubMed] [Google Scholar]
- Tinberg C. E.; Song W. J.; Izzo V.; Lippard S. J. Biochemistry 2011, 50, 1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinberg C. E.; Lippard S. J. Acc. Chem. Res. 2011, 44, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


