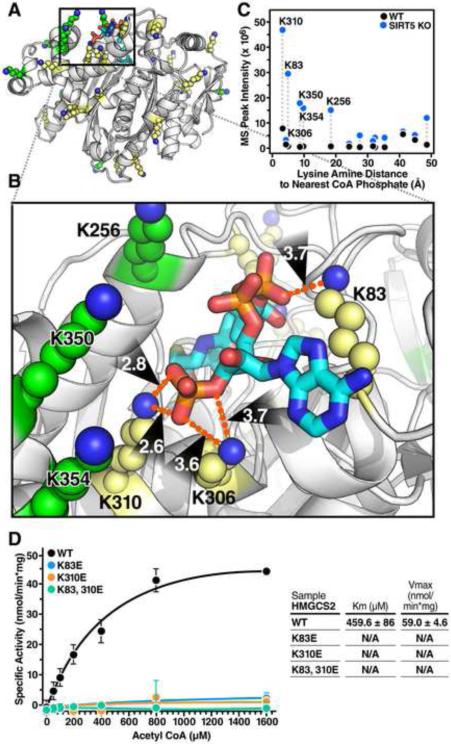
Figure 7. HMGCS2 mutations at sites of succinylation within the substrate binding region regulate its enzymatic activity.
(A) Ribbon diagram of the crystal structure of human HMGCS2 (PDB entry 2WYA) bound with HMG-CoA. Lysines fully observed in the crystal structure are shown in yellow, and lysines without fully defined conformations in the crystal structure are modeled in PyMol using low-energy all-trans rotamers as allowed by local backbone and steric interactions and displayed as green atomic spheres at 50% scale. (B) The interactions between several lysine side chains and the negatively charged CoA phosphate groups in the substrate binding pocket are shown in detail. Lysine amine nitrogens for residues 83, 306 and 310 are within 4Å of the phosphate oxygen atoms. (C) The observed mass spectrum peak intensity of succinyl-lysine-containing peptides plotted with respect to their distance to the nearest CoA phosphate. Dotted lines connect the same peptide identified in WT and Sirt5−/− mice. (D) Steady state kinetic analysis of WT HMGCS2 and succinyllysine mimetics HMGCS2-K83E, HMGCS2-K310E, and HMGCS2-K83,310E enzymatic activity as measured by DTNB detection of CoA-SH released from increasing concentrations of acetyl-CoA at 412 nM. Graph is representative of two independent experiments, n=3 measurements/sample, mean ± SD. Inset contains values for average Km and Vmax ± SEM. The purity and equal amounts of immunoprecipitated HMGCS2 WT or mutants are shown in Figure S5.