Summary
The exquisite control of growth factor function by heparan sulfate (HS) is dictated by the tremendous structural heterogeneity of sulfated modifications. It is not known how specific HS structures control growth factor-dependent progenitor expansion during organogenesis. We isolated KIT+ progenitors from fetal salivary glands during a stage of rapid progenitor expansion and profiled HS biosynthetic enzyme expression. Enzymes generating a specific type of 3-O-sulfated-HS (3-O-HS) are enriched, and FGF10/FGFR2b signaling directly regulates their expression. Bioengineered 3-O-HS binds FGFR2b and stabilizes FGF10/FGFR2b complexes in a receptor- and growth factor-specific manner. Rapid autocrine feedback increases 3-O-HS, KIT and progenitor expansion. Knockdown of multiple Hs3st isoforms limits fetal progenitor expansion, but is rescued with bioengineered 3-O-HS, which also increases adult progenitor expansion. Rapidly altering a specific 3-O-sulfated epitope provides a cellular mechanism to modulate the response to FGFR2b signaling and control progenitor expansion. 3-O-HS may expand KIT+ progenitors in vitro for regenerative therapy.
Keywords: Heparan sulfate 3-O-sulfotransferase, FGF10, heparan sulfate, KIT, FGFR2b, salivary gland development, SMG, progenitor proliferation, progenitor expansion
INTRODUCTION
During organogenesis, fibroblast growth factor receptor 2b (FGFR2b) signaling is essential for progenitor survival and proliferation in the submandibular gland (SMG), lung, pancreas, tooth and skin (De Moerlooze et al., 2000; Petiot et al., 2003). It is well established that heparan sulfate proteoglycans are coreceptors for FGFR2b (Mohammadi et al., 2005) and specific HS structures are critical for FGFR2b-dependent proliferation (reviewed in Patel and Hoffman, 2013; Patel et al., 2006). The genetic deletion of HS biosynthetic enzymes highlights that HS is essential for organogenesis (Bishop et al., 2007). Due to the tremendous variation in sulfate modifications of HS, the sulfotransferase enzymes provide an attractive mechanism to increase the cellular specificity to HS-binding growth factors and to fine-tune the biological responses to FGFR2b signaling.
The HS 3-O-sulfotransferase (Hs3st) family is the largest family of sulfotransferase enzymes with seven isoforms in mice and humans. However, 3-O-sulfation is the least abundant sulfate modification and occurs at the terminal step of HS biosynthesis within the Golgi. Although all seven isoforms add a 3-O-sulfate, they generate two types of modifications (Lawrence et al., 2007; Xu et al., 2008). The first, termed Hs3st1-like activity, occurs when Hs3st1 adds a sulfate to a glucosamine adjacent to either a glucuronic or an iduronic acid in the HS chain. This well-studied type of 3-O-sulfated modification occurs in the antithrombin (AT) binding pentasaccharide (Lindahl et al., 1980). The second, termed Hs3st3-like activity, occurs when Hs3st isoforms -2, -3a1, -3b1, -4, or -6 add a 3-O-sulfate to a glucosamine adjacent to a 2-O-sulfated iduronic acid. For example, this forms a cell surface HS epitope that allows herpes simplex virus (HSV-1) entry (Shukla et al., 1999). Lastly, Hs3st5 can generate both types of modifications (Liu and Pedersen, 2007).
Only two of the Hs3st’s have been genetically deleted in mice. Loss of Hs3st1 results in postnatal lethality with intrauterine growth retardation (HajMohammadi et al., 2003) and mice deficient in Hs3st2 are viable (Hasegawa and Wang, 2008). Hs3st’s have distinct roles in other organisms. For instance, a reduction of Hs3st-B affects D. melanogaster neurogenesis (Kamimura et al., 2004), hst-3.1 and hst-3.2 mediate neurite branching in C. elegans, (Tecle et al., 2013), while in D. rerio Hs3st5 controls cilia length, Hs3st6 regulates cell motility (Neugebauer et al., 2013) and Hs3st7 regulates cardiac contraction (Samson et al., 2013). However, the specific roles of the Hs3st3 isoforms during mammalian development are unknown.
Recently, we reported that the highly proliferative distal endbuds of SMGs and lungs contain progenitors expressing the KIT receptor and FGFR2b, and that combined KIT/FGFR2b signaling is critical for their maintenance and expansion (Lombaert et al., 2013). Here we discover that KIT+ epithelial progenitors preferentially synthesize Hs3st3-modified HS to rapidly increase FGFR2b-mediated signaling and proliferation. 3-O-HS-dependent signaling results in autocrine feedback to upregulate Hs3st3 and Kit expression. We propose that 3-O-HS controls the FGFR2b-dependent expansion of KIT+ progenitors during organogenesis and during the in vitro expansion of adult progenitors.
RESULTS
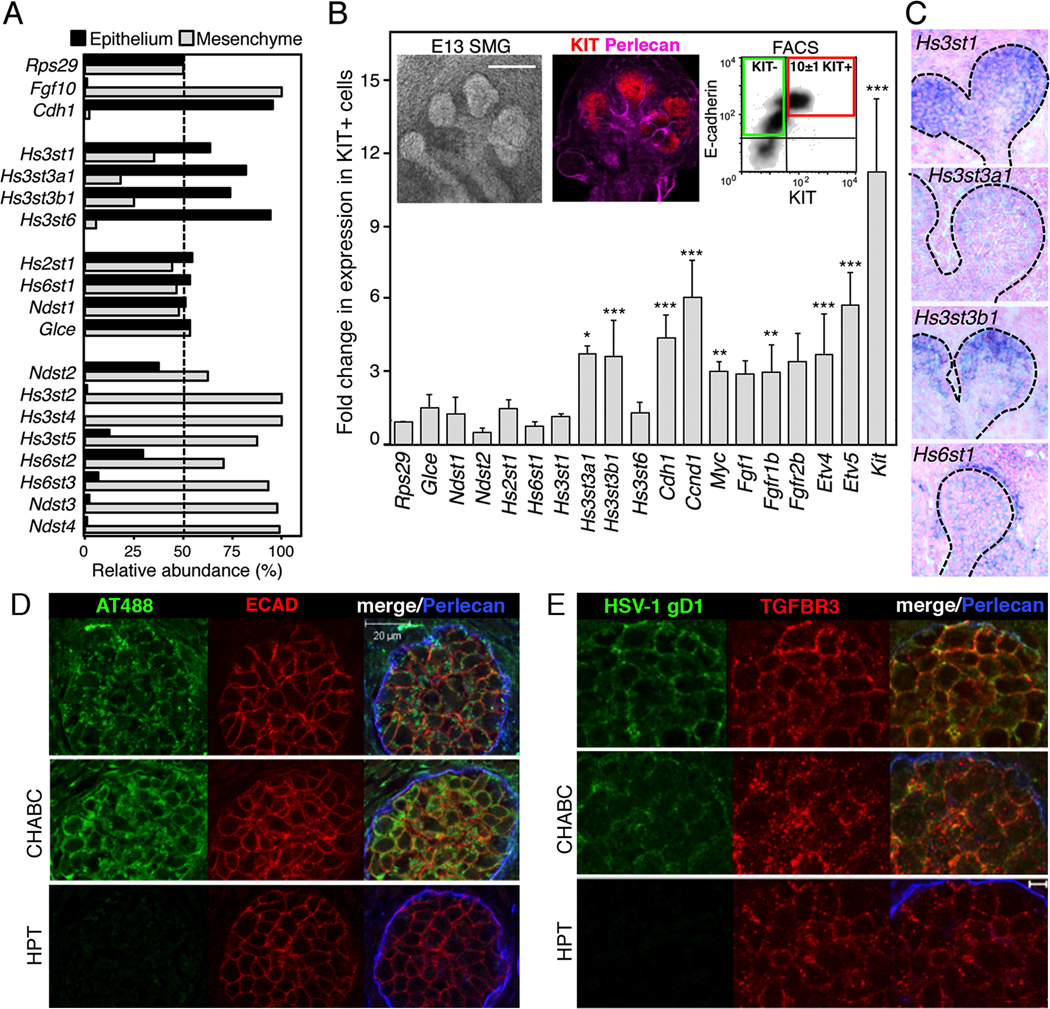
KIT+ epithelial cells are enriched in Hs3st3a1 and Hs3st3b1 transcripts and 3-O-sulfated HS is localized in KIT+ epithelial endbuds
Since HS is required for high-affinity binding of FGF10 to FGFR2b, we hypothesized that a progenitor-specific HS modification on proteoglycans controlled FGFR2b-dependent proliferation. We first screened the HS biosynthetic enzyme expression in SMG epithelium and mesenchyme at embryonic day (E) 13, to identify which enzymes were expressed in the epithelium (Figure 1A). Transcripts for Hs3st1, Hs3st3a1, Hs3st3b1 and Hs3st6 were predominantly in epithelia, whereas Hs3st2, Hs3st4 and Hs3st5 were in the mesenchyme. Other HS biosynthetic enzymes such as Hs2st1, Hs6st1 and Ndst1 were equally expressed in both compartments. Cdh1 and Fgf10 served as controls for the separation of the epithelium and mesenchyme, respectively. We then used KIT and E-cadherin (ECAD) antibodies to FACS sort the epithelial KIT+ progenitors, which are mainly localized in the endbuds in E13 SMGS (Figure 1B). We profiled the expression of HS biosynthetic enzymes in the KIT+ECAD+ and KIT−ECAD+ cell populations using qPCR. Interestingly, the FACS sorted KIT+ cells showed increased expression of specific HS-3-O-sulfotransferase isoforms, Hs3st3a1 and Hs3st3b1, compared to KIT− cells. We confirmed that KIT+ cells are enriched for genes associated with proliferation and FGFR signaling by the increase in Ccnd1, Fgfr1b, and the downstream targets of FGFR signaling Etv4, Etv5, and Myc (Figure 1B). We screened HS proteoglycan core proteins in the KIT+ cells, although some were enriched, none were significantly increased in expression (Figure S1A). These data indicate that the highly proliferating KIT+ECAD+ progenitors express a unique set of sulfotransferase enzymes.
Figure 1. KIT+ epithelial endbud progenitors are enriched for HS 3-O-sulfotransferase expression and 3-O-HS is localized in the epithelial endbuds.
(A) qPCR analysis of isolated E13 SMG epithelia and mesenchyme. Gene expression was normalized to Rps29.
(B) The expression levels of HS biosynthetic enzymes were compared by qPCR in cDNA from the KIT+ versus KIT− ECAD+ FACS sorted cells. Gene expression was normalized to expression in the KIT− cells and Rps29. A false discovery rate (Q) for multiple unpaired t-tests was set to 5%. *p ≤ 0.05 **p ≤ 0.01 and ***p ≤ 0.001. Error bars: SEM. Images are single confocal projection with KIT (red) and Perlecan (pink) labeling in an E13 SMG. Scale bar; 200 µm. FACS plot shows KIT+ ECAD+ cells (red box) and KIT− ECAD+ cells (green box) sorted from E13 SMGs.
(C) Sections of whole-mount in situ analysis of E13 SMG confirm the localization of Hs3st isoforms in the endbud (dotted line); counterstained with nuclear fast red.
(D) Staining of E13 SMGs with labeled antithrombin (AT488, green). Heparitinase (HPT) but not chondroitinase (CHABC) treatment for 2 hr abolished AT488 labeling. Perlecan (blue) localized in the basement membrane and ECAD (red) labeled the epithelium. Images are single confocal sections, scale bar: 20 µm.
(E) Staining with HSV-1 gD285 protein localized Hs3st3-type 3-O-HS at the epithelial cell surface. Heparitinase treatment also decreased binding of HSV-1 gD (green). Perlecan (blue) and TGFBR3 (red), which stains the epithelium, are shown. Images are single confocal sections, scale bar: 5 µm.
See also Figure S1.
Whole-mount in situ analysis of E13 SMGs confirmed the localization of Hs3st1, Hs3st3a1 and Hs3st3b1 in epithelial endbuds, and Hs6st1 in both the epithelium and mesenchyme (Figure 1C and S1B). Hs3st6 was barely detectable by qPCR and we were unable to detect it by in situ analysis (data not shown). We also analyzed the temporal expression of the Hs3st enzymes throughout SMG development by qPCR (Figure S1C). Hs3st1, Hs3st3a1 and Hs3st3b1 were more abundant early in SMG development when KIT+ progenitors proliferate to increase organ size. By comparing the expression with other fetal organs at E12, including the heart, kidney, liver, limb, lung and brain (Figure S1D), distinct tissue-specific expression patterns of 3-O-sulfotransferases were apparent. For example, Hs3st4 and Hs3st5 were highly expressed in the brain, whereas Hs3st6 was high in the kidney (Figure S1D).
Next, it was important to confirm that 3-O-HS was present on SMG epithelial cells (Figure 1D and E). We incubated SMGs with fluorescent Alexa 488-labeled antithrombin (AT488), which recognizes the 3-O-HS pentasaccharide epitope generated by the Hs3st1 enzyme (Girardin et al., 2005). We also incubated SMGs with a truncated ectodomain of HSV-1gD, which binds to the Hs3st3-modified HS (Krummenacher et al., 1999; Shukla et al., 1999). Both AT488 and HSV-1gD specifically labeled HS on the SMG epithelial cell surface. Pretreatment with heparitinase III (HPT) specifically abolished labeling, whereas chondroitinase ABC (CHABC), which removes chondroitin sulfates, did not (Figure 1D and E). In addition, incubation with excess AT-binding pentasaccharide or unlabeled AT competed the AT488 binding (not shown). Taken together, we show that Hs3st enzymes are specifically localized to epithelial endbuds that contain the KIT+ progenitors, and that 3-O-HS is present on their cell surface.
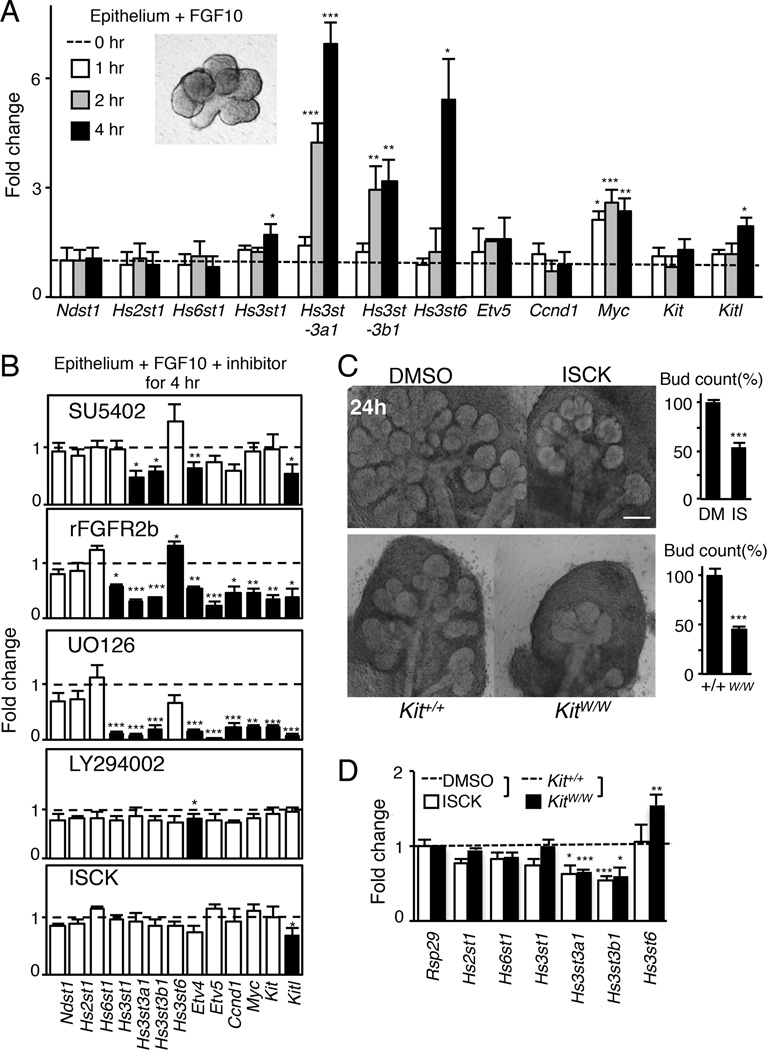
Hs3st3 expression is directly regulated by FGFR2b signaling
We then investigated whether either FGF10/FGFR2b or KIT signaling directly regulates the expression of Hs3st isoforms. Isolated epithelia were treated with FGF10 for 1, 2, and 4 hours (hr), and Hs3st isoform expression was measured by qPCR. No morphological differences in the treated epithelium were observed at these short time points (Figure 2A). Surprisingly, Hs3st3a1 and Hs3st3b1 expression increased by 2 hr and by 4 hr Hs3st3a1 expression increased 7-fold. By 4 hr, the expression of Hs3st1 and Hs3st6 had also increased. As expected, Myc, an early response gene of FGF10/FGFR2b signaling, increased within 1 hr while the expression of Ndst1, Hs2st1 and Hs6st1 did not change during this time (Figure 2A). We also confirmed that the expression of Kitl increased at 4 hr whereas Kit did not, as previously shown (Lombaert et al., 2013). Importantly, the increase in 3-O-sulfotransferase genes occurred more rapidly than Etv5, a known downstream target of FGF10/FGFR2b signaling.
Figure 2. Hs3st expression is rapidly regulated by FGFR2b-dependent MAPK signaling and is reduced when KIT signaling is reduced.
(A) Isolated E13 epithelia (insert image) were treated with 400 ng/ml FGF10 for 1, 2 or 4 hr, which does not affect morphogenesis. Gene expression was normalized to epithelium at 0 hr and Rps29.
(B) SMGs (E13) were treated for 4 hr with an FGFR inhibitor (SU5402, 5µM), recombinant FGFR2b (25nM), a MAPK inhibitor (UO126, 20µM), a PI3-K inhibitor (LY294002, 25µM) and a SCF/KIT inhibitor (ISCK03, 20µM). qPCR analysis was normalized to DMSO or rFGFR1b (a control for rFGFR2b). No morphological changes were observed between treatments after 4 hr (insert image).
(C) Inhibition of KIT signaling in SMGs with a KIT inhibitor (ISCK03, 20µM) or in vivo using E14 KitW/W mice reduced SMG branching morphogenesis compared to controls. The bud count was normalized (%) to control. Scale bar; 200 µm.
(D) SMGs (E13) either treated with a KIT inhibitor (ISCK03, 20µM) for 24 hr or isolated from E13 KitW/W mice have reduced expression of Hs3st3a1 and Hs3st3b1. Gene expression was normalized to DMSO-carrier control or wild-type control and Rps29.
Error bars: SEM. ANOVA (A–C); Student t-test (D); * p < 0.05, ** p < 0.01 and *** p < 0.001.
To identify the downstream signaling pathway regulating Hs3st expression, isolated epithelia were treated for 4 hr with FGF10 and either an FGFR inhibitor (SU5402), recombinant FGFR2b, a MAPK inhibitor (UO126), a PI3-K inhibitor (LY294002) or a KIT inhibitor (ISCK) (Figure 2B). As predicted, no morphological differences were observed, but there was reduced expression of Hs3st3a1 and Hs3st3b1 in epithelia treated with the FGFR inhibitor, recombinant FGFR2b, and MAPK inhibitor, but not with the PI3-K or KIT inhibitors (Figure 2B). There was also reduced expression of Etv4, Etv5, Ccnd1, Myc, Kit and Kitl with recombinant FGFR2b and MAPK inhibitors, and reduced expression of Etv4 with the PI3-K and FGFR inhibitors. The specific KIT inhibitor did not affect the expression of any genes except for Kitl at 4 hr.
Although KIT signaling did not directly regulate Hs3st3 expression, we previously reported that a reduction in KIT signaling in vivo diminishes organ growth by depleting KIT+ progenitors (Lombaert et al., 2013). Therefore, we analyzed the expression of Hs3st3a1 and Hs3st3b1 in E13 SMGs from KitW/W mice, which have a mutation in KIT that inhibits signaling, as well as SMGs treated with the KIT inhibitor for 24 hr. In both situations, there was a decrease in branching morphogenesis (Figure 2C) and reduced expression of Hs3st3a1 and Hs3st3b1 (Figure 2D). Interestingly, Hs3st6 expression increased in KitW/W SMGs compared to the wild-type glands, suggesting that in vivo compensation with other Hs3st isoforms may occur. Overall, these data suggest that an FGF10/FGFR2b-mediated MAPK pathway rapidly and directly regulates Hs3st3 expression, and that the reduced proliferation by loss of KIT signaling correlates with reduced levels of Hs3st3s.
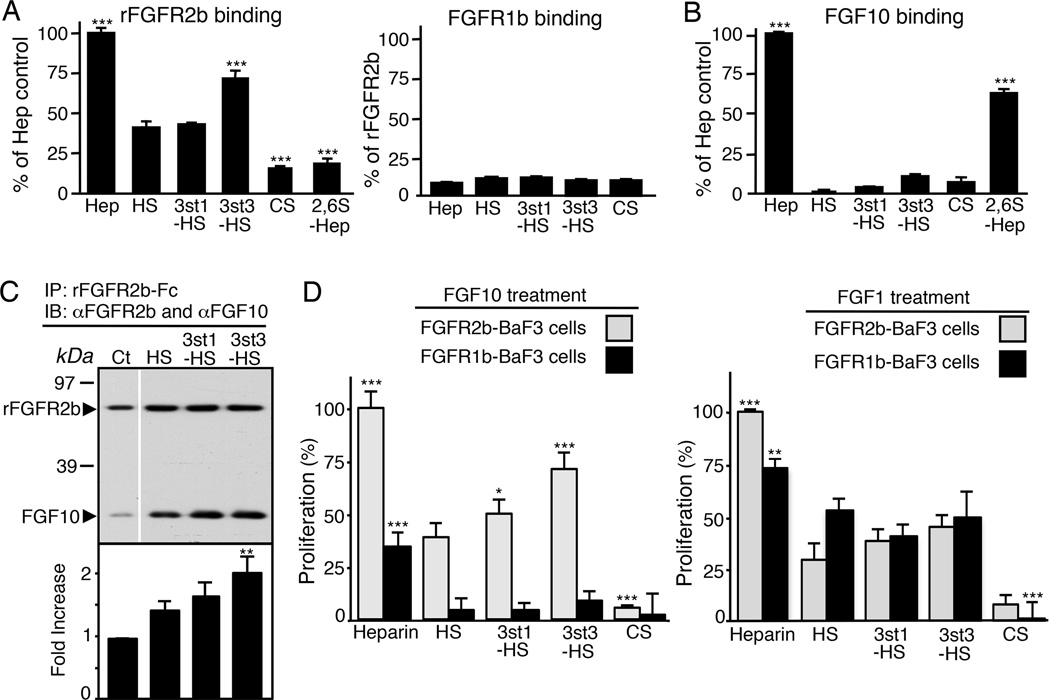
3-O-sulfated-HS binds FGFR2b to enhance 3-O-HS/FGF10/FGFR2b ternary complex formation and proliferation
In order to investigate the molecular mechanism of how 3-O-HS could rapidly control progenitor expansion, we bioengineered HS with 3-O-sulfated epitopes generated by different Hs3st enzymes. We used HS isolated from bovine kidney, because it has low levels of endogenous 3-O-sulfation (Xu et al., 2008). This kidney HS (simply termed HS here) was treated with recombinant Hs3st1 and Hs3st3 enzymes to generate two types of 3-O-HS, denoted as 3st1-HS and 3st3-HS, respectively (Figure S2A). The enzymatic treatment was monitored by radioactive sulfate incorporation and only one or two 3-O-sulfates were added per 14 kDa HS chain, which had on average 48 sulfates/chain, thereby increasing overall sulfation by only 2–4% (see Experimental procedures and (Xu et al., 2008)). The current model of an HS/FGF10/FGFR2b ternary signaling complex predicts that HS could function to promote FGFR2b dimerization, stabilize the interactions of FGF10 with the receptor and/or stabilize direct receptor-receptor contacts (Mohammadi et al., 2005). We used a variety of biochemical assays to define how 3-O-HS functions.
To determine whether 3-O-HS could bind to either FGFR2b or FGF10, a solid-phase HS-binding assay was used. The 3-O-HS was immobilized in GAG-binding plates and recombinant FGFR2b-Fc or FGF10 were added. Binding was measured with an anti-Fc antibody to detect FGFR2b or an anti-FGF10 antibody. Heparin, which binds both FGFR2b and FGF10 and is highly sulfated (including 3-O-sulfation), was used as a positive control and chondroitin sulfate A as a negative control. We observed increased FGFR2b binding to 3st3-HS compared to 3st1-HS or HS (Figure 3A). As expected, recombinant FGFR1b did not bind to HS (Figure 3A). FGF10 also did not bind to the immobilized 3-O-HS any more than chondroitin sulfate (Figure 3B), even at concentrations 10-fold higher than that used for FGFR2b binding. Heparin decasaccharides (2,6S-Hep) that increase FGF10 bioactivity (Patel et al., 2008) were used as a positive control for FGF10 binding and bound more FGF10 than FGFR2b (Figure 3B). These data suggest that 3-O-HS preferentially interacts with FGFR2b and not FGF10.
Figure 3. 3st3-HS preferentially binds FGFR2b to increase ternary complex formation of FGF10/FGFR2b, and proliferation of FGFR2b-expressing BaF3 cells.
(A–B) Binding of FGFR2b and FGFR1b (A) or FGF10 (B) to immobilized GAGS. FGFR2b, but not FGF10, specifically binds to immobilized 3st3-HS. ELISA assays were performed in triplicate and repeated at least three times. Error bars: SEM.
(C) 3-O-HS increases the amount of FGF10 immunoprecipitated with FGFR2b. The intensity of the protein bands was normalized to FGFR2b and shown as a fold change in ratio of FGF10/FGFR2b. The graphs are 3 experiments combined and the blots from a representative experiment.
(D) BaF3 cells expressing FGFR2b or FGFR1b were incubated with 25nM GAGS (CSA, heparin, HS, 3st1-HS, or 3st3-HS) and 50nM FGF10 or 5nM FGF1. After 40 hr, the relative cell proliferation (A490) was determined. Data were normalized to FGFR2b-BaF3 cells treated with FGF10 or FGF1 and heparin.
Error bars: SEM. ANOVA; * p < 0.05, ** p < 0.01 and *** p < 0.001.
See also Figure S2.
To determine whether 3-O-HS stabilizes FGF10/FGFR2b complex formation, equimolar amounts of recombinant FGFR2b, FGF10 and HS were mixed, and then FGFR2b was immunoprecipitated and blotted for both FGFR2b and FGF10. As expected, HS increased the ternary complex formation and more FGF10 was detected, compared to when HS was not added. However, the addition of either 3st1-HS or 3st3-HS further increased the amount of FGF10 pulled down in the complex (Figure 3C). This shows that 3-O-HS enhances the ternary 3-O-HS/FGF10/FGFR2b signaling complex formation by binding to FGFR2b, which may affect downstream outcomes.
To determine whether 3-O-HS specifically enhances FGFR2b-dependent proliferation, we used BaF3 cell assays. These cells do not express endogenous HS or FGFRs, and they do not proliferate in response to FGF or heparin alone (Zhang et al., 2006). Proliferation of FGFR2b- and FGFR1b-expressing BaF3 cells was compared following incubation with either FGF10 or FGF1 and 3-O-HS, using heparin and chondroitin sulfate as positive and negative controls, respectively. With FGF10 treatment, both 3st1-HS and 3st3-HS increased FGFR2b-BaF3 but not FGFR1b-BaF3 cell proliferation compared to HS, with 3st3-HS having the greatest increase (70% of heparin) (Figure 3D). With FGF1 treatment, the proliferation of FGFR1b-BaF3, FGFR2b-BaF3 and FGFR3c-BaF3 cells was similar with either 3-O-HS or HS (Figure 3D and Figure S2), suggesting the response to 3-O-HS is both receptor- and growth factor-dependent.
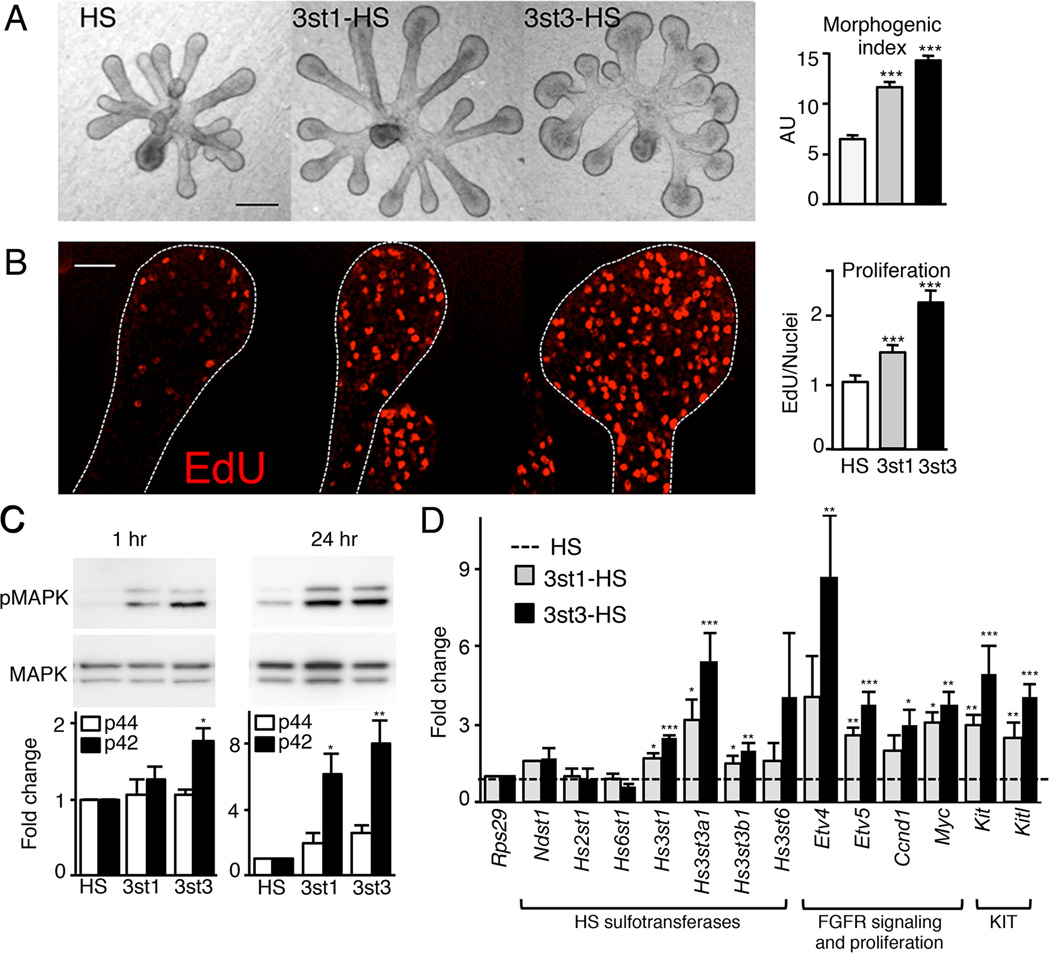
3-O-HS/FGF10/FGFR2b increases SMG epithelial cell proliferation and regulates FGFR2b-dependent gene expression
To confirm that 3-O-HS also elicits an FGF10-dependent mitotic response in SMG epithelium, we measured changes in epithelial morphogenesis and proliferation after the addition of either 3st1-HS or 3st3-HS compared to HS. The addition of 3st3-HS increased epithelial morphogenesis more than 3st1-HS (Figure 4A). Morphogenesis was expressed as a morphogenic index (endbud number × width × duct length, in AU) (Figure 4A). The addition of 3st3-HS also had a greater effect (2-fold compared to HS) than 3st1-HS on proliferation, which was quantified using EdU (5’-ethynyl-2’-deoxyuridine) labeling (Figure 4B). To confirm that this proliferation was MAPK-dependent (as suggested in Figure 2), we measured p42/44MAPK downstream of 3-O-HS after 1 hr and 24 hr. Both 3-O-HS increased p42/44MAPK compared to HS within 1 hr, and this was maintained at 24 hr. Again, the increase with 3st3-HS was greater than with 3st1-HS (Figure 4C).
Figure 4. 3-O-sulfated-HS increases FGF10-dependent epithelial morphogenesis by increasing proliferation and FGFR2b-downstream gene expression.
(A) FGF10 cultured E13 SMG epithelia treated with HS, 3st1-HS or 3st3-HS for 28 hr. 3st3-HS increased endbud morphogenesis of FGF10-treated epithelium compared to HS (control). Morphogenic index (endbud number × width × duct length, in AU) increases with addition of 3-O-HS. Scale bar; 50 µm.
(B) Single confocal sections showing cell proliferation (red) in epithelia cultured for 28 hr. Quantification of fluorescence intensity normalized to total nuclei staining and expressed as a ratio to the HS treated epithelia. At least five epithelia/condition were used for quantification from three independent experiments. Scale bar; 10 µm.
(C) 3st3-HS increased MAPK signaling within 1 hr and both 3st-HS maintained it at 24 hr in FGF10-cultured epithelia. Band intensity of pMAPK was normalized to total MAPK and expressed as a fold change in ratio compared to HS. The graphs are 3 experiments combined and the blots from a representative experiment.
(D) Gene expression was measured by qPCR and normalized to epithelia cultured for 28 hr with HS and Rps29.
Error bars: SEM. ANOVA; *p < 0.05; **p < 0.01, and ***p < 0.001.
See also Figure S3.
We then measured gene expression downstream of 3-O-HS treatment. Both 3st1-HS and 3st3-HS specifically increased Hs3st1, Hs3st3a1 and Hs3st3b1, but not the other HS sulfotransferases (Figure 4D), suggesting that positive autocrine feedback increased 3-O-HS. In addition, 3-O-HS increased the expression of genes downstream of FGFR2b signaling Etv4, Etv5, Myc, and Ccnd1, and Kit and Kitl. Although 3st1-HS and 3st3-HS affected similar genes, 3st3-HS had a greater effect on expression. To confirm that this occurs in other systems, we treated E11 lung endbuds with FGF10 and 3-O-HS or HS (Figure S3). Similar to SMGs, the 3st3-HS increased both morphogenesis and the expression of FGFR2b-dependent transcription factors involved in lung endbud progenitor development, Etv5, Sox9 and Id2 (Lombaert et al., 2013; Rock and Hogan, 2011). Taken together, these data suggest that 3-O-HS increases and maintains FGFR2b-dependent signaling and proliferation of endbud progenitors.
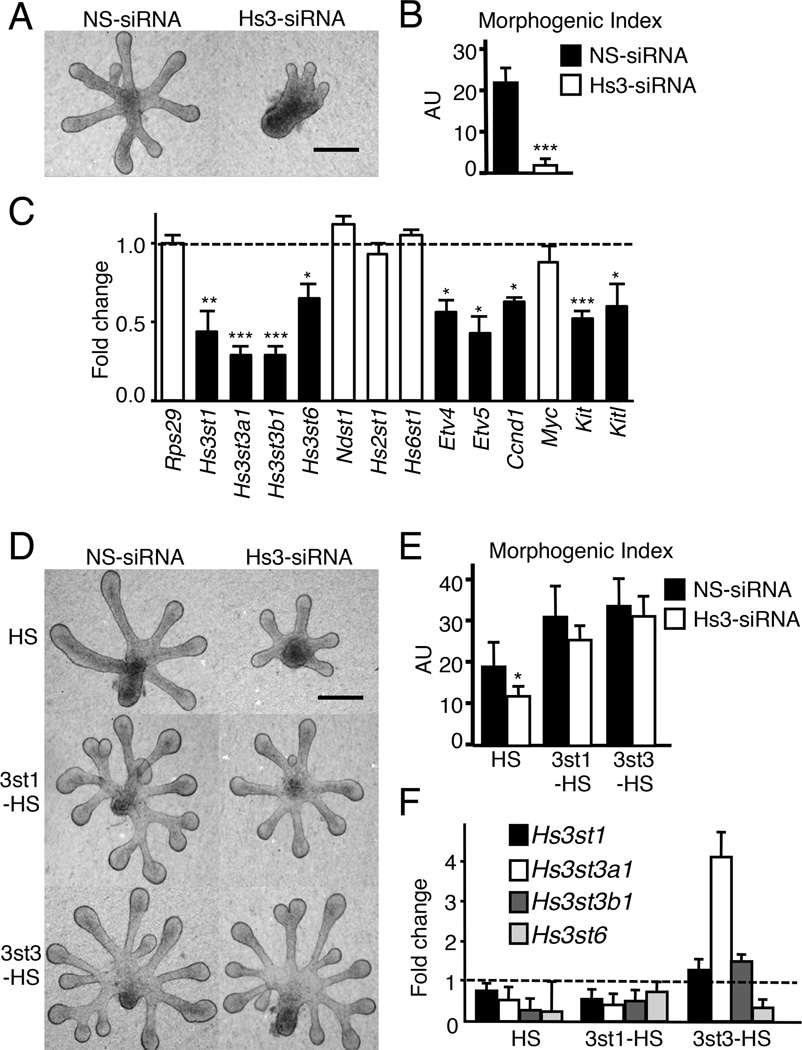
Knockdown of Hs3sts reduces FGF10/FGFR2b-dependent growth and both are rescued with exogenous 3-O-HS
Next, we predicted that reducing Hs3st gene expression by RNAi would reduce FGFR2b-dependent epithelial proliferation. We used at least two different siRNAs for each isoform but the knockdown of individual Hs3st3 isoforms or Hs3st1 did not affect morphogenesis (Data not shown). Therefore, we designed an siRNA that targeted common regions of the Hs3st3a1, Hs3st3b1 and Hs3st6 isoforms and pooled it with one targeting Hs3st1. Together they reduced epithelial morphogenesis and Hs3st expression (Figure 5A and 5B). The siRNAs reduced Hs3st1, Hs3st3a1, Hs3st3b1 and Hs3st6 but not the other HS sulfotransferases such as Ndst1, Hs2st1 or Hs6st1 (Figure 5C). Hs3st knockdown also reduced expression of the Etv4, Etv5, Ccnd1, Kit and Kitl (Figure 5C), consistent with 3-O-HS being involved in FGF10/FGFR2b signaling and proliferation of KIT+ progenitors. We predicted that exogenous 3-O-HS would rescue the effects of knocking down Hs3st expression and increase FGFR2b-dependent proliferation. We added HS to control siRNA-treated epithelium at a dose that did not increase proliferation (Figure 5D and E). However, similar concentrations of both 3st1-HS and 3st3-HS rescued morphogenesis in Hs3st-siRNA treated epithelium compared to HS alone. Despite similar concentrations, 3st3-HS had greater effects (Figure 5D and 5E) and also overcame the effects of siRNA and increased the expression of Hs3st1, Hs3st3a1 and Hs3st3b1 (Figure 5F). Together, these data show that 3-O sulfated HS controls FGF10/FGFR2b-dependent proliferation, and that exogenous 3-O-HS increases Hs3st gene expression to overcome the effects of the siRNA in culture. Overall, our data suggested that 3st3-HS might be a tool to specifically increase FGFR2b-dependent proliferation and expand KIT+ progenitors.
Figure 5. Knockdown of Hs3sts decreases epithelial morphogenesis and Hs3st expression, which are both rescued by exogenous 3st3-HS.
(A) Light micrographs of FGF10-cultured E12 SMG epithelia were treated for 40 hr with non-silencing (NS) or Hs3st siRNA. Scale bar; 50 µm.
(B) Quantification of epithelial morphogenesis after 40 hr of siRNA transfection. Knockdown of the Hs3sts inhibited branching compared to NS-siRNA. Error bars SEM. ANOVA; ***p<0.001, compared to control.
(C) Knockdown of Hs3sts decreased expression of Etv4, Etv5, Ccnd1, Myc, Kit and Kitl. Gene expression was measured by qPCR and normalized to NS-siRNA (dotted line). Error bars: SEM. Student’s t-test *p < 0.05; **p < 0.01, and ***p < 0.001.
(D) Light micrographs of FGF10-cultured E12 SMG epithelia were treated with NS or Hs3st siRNA. Exogenous HS, 3st1-HS, or 3st3-HS was added to these cultures following 16 hr of siRNA-treatment. Scale bar; 50 µm.
(E) Quantification of epithelial morphogenesis after 40 hr of siRNA transfection. Error bars: SEM. ANOVA; ***p<0.001, compared to epithelia treated with NS and HS.
(F) Gene expression of Hs3st1, Hs3st3a1 and Hs3st3b1 is partially restored by 3st1-HS and 3st3-HS. Gene expression was measured by qPCR and normalized to NS-siRNA (dotted line). Error bars: SEM.
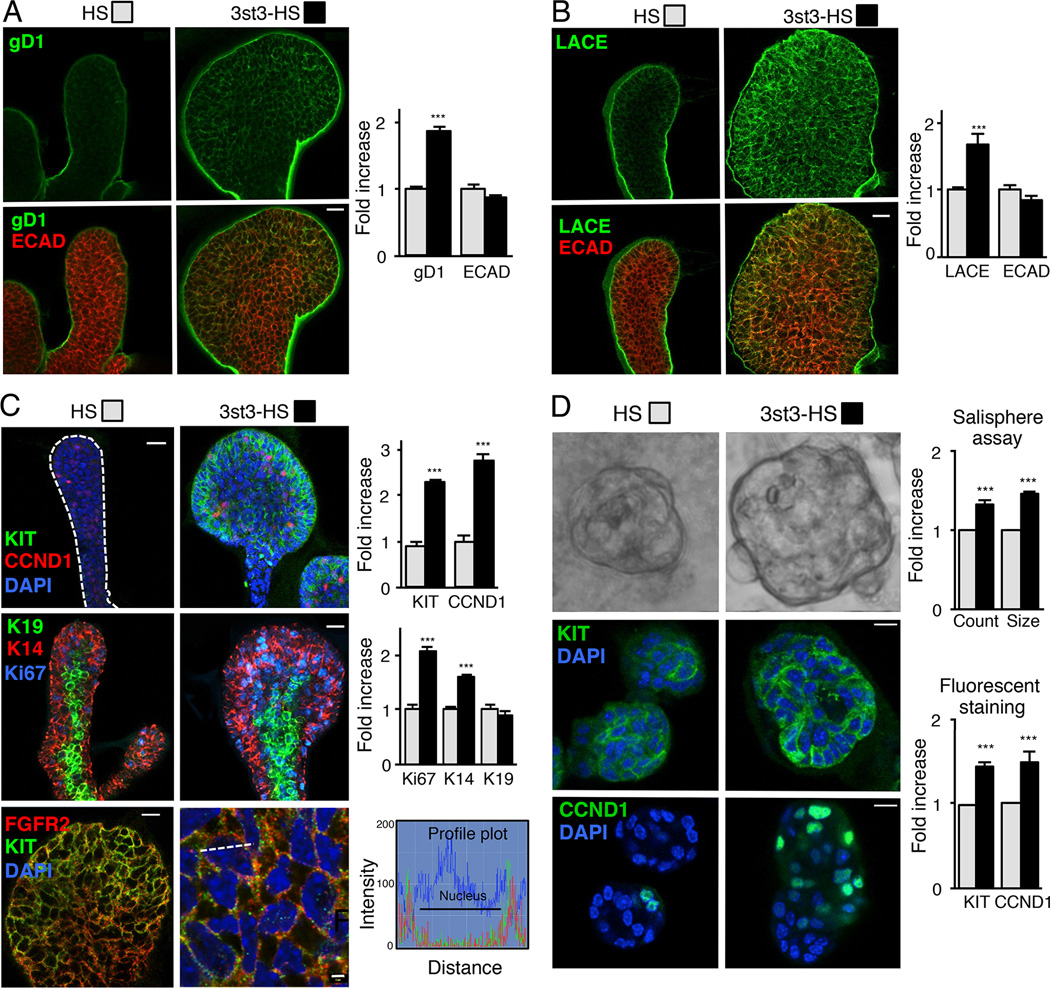
3st3-HS increases endogenous 3-O-sulfated HS and increases FGF10/FGFR2b binding to the epithelium
We next confirmed that treatment with 3st3-HS also increased the endogenous 3-O-sulfated HS on the epithelium by staining with gD1. An increase in gD1 binding was detected on epithelium treated with 3st3-HS compared to control HS (Figure 6A). In addition, a Ligand and Carbohydrate Engagement (LACE) assay was performed with recombinant FGF10 and FGFR2b-Fc to determine whether the increase in endogenous gD1-binding HS also resulted in increased binding of the FGF10/FGFR2b complex, which was detected with anti-human-Fc antibodies. Indeed, an increase in FGF10/FGFR2b assembly was detected on epithelia treated with 3st3-HS compared to HS (Figure 6B). These data are consistent with the hypothesis that an increase in endogenous 3-O-sulfated HS increases FGF10/FGFR2b signaling.
Figure 6. 3-O-sulfated-HS increases endogenous 3-O-HS, FGF10/FGFR2b-Fc binding, proliferation and KIT expression.
(A–C) FGF10 cultured E13 SMG epithelia treated with HS or 3st3-HS for 28 hr were stained. (A) gD1 staining (green) or (B) with FGF10/FGFR2b-Fc (LACE assay, green) and ECAD (red). (C) CCND1, KIT (green) and DAPI (blue) top panels, K14 (red), K19 (green), Ki67 (blue) middle panels, and FGFR2 (red), KIT (green) and DAPI (blue) lower panels. The profile plots (lower right) quantify the relative intensity of the KIT and FGFR2 protein distribution at the position along the white dotted line in the overlay image. The fluorescence signal from anti-KIT-Cy2 colocalized with anti-FGFR2-Cy3 immunofluorescence signal with an overlap coefficient of r=0.54 ± 0.015. Images are 2 µm confocal sections. Scale bars: 10 µm and 2 µm in lower right panel. Quantification of fluorescence intensity normalized to total nuclei staining and expressed fold increase compared to HS treated epithelia. At least five epithelia from three independent experiments were used for quantification.
(D) 3-O-HS treatment increases salisphere size (upper panel). Quantification of the number and diameter of salispheres formed after 3 days. 3-O-HS increases both the count and size of salispheres compared to HS. Confocal sections of salispheres cultured for 6 days with HS or 3st3-HS stained with KIT (green, middle panel) and CCND1 (green, lower panel). Quantification of KIT and CCND1 fluorescence intensity normalized to total nuclei staining and expressed as a fold increase compared to HS treated salispheres. At least five salispheres from each of three independent experiments were used for quantification. Scale bar; 10 µm.
Error bars: SEM. Student’s t-test; *p < 0.05, **p < 0.01, and ***p < 0.001.
3st3-HS specifically expands fetal and adult epithelial KIT+FGFR2b+ progenitors
Since combined KIT/FGFR2b signaling increases the expansion of KIT+FGFR2b+K14+ epithelial endbud progenitors (Lombaert et al., 2013), we predicted that exogenous 3st3-HS would specifically enhance their expansion. We immunostained epithelia treated with 3st3-HS or HS for KIT, CCND1, Ki67, K14, and K19 (Figure 6C). 3st3-HS-treatment increased staining compared to HS, for KIT, K14 and the proliferative markers CCND1 and Ki67 (Figure 6C top and middle panels). 3st3-HS did not affect staining of K19, a marker of ductal cells (Figure 6C middle panels). We further confirmed that 3st3-HS-treated epithelial KIT+ progenitors expressed FGFR2b (Figure 6C, lower panels). A profile plot shows that both KIT and FGFR2b localize near the membrane at a calculated overlap coefficient of r=0.54 ± 0.015.
Since KIT+FGFR2b+ progenitors are present in the adult SMG, we asked whether 3-O-HS expanded them in vitro, which is an important step to obtain more progenitors for transplantation. Thus, we used in vitro spheroid culture of adult SMG progenitors, termed salispheres, as previously used to enrich for KIT+ progenitors for autologous transplantation to regenerate irradiation-damaged salivary glands (Lombaert et al., 2008). The addition of 3st3-HS and FGF10 increased the number and size of salispheres by day 3, an indicator of increased progenitor numbers, compared to HS treated, (Figure 6D). Remarkably, even at 6 days of culture, the salispheres contained cells expressing KIT that were actively proliferating (CCND1+) (Figure 6D). Together, these data indicate that exogenous 3st3-HS expands and prolongs the proliferation of adult KIT+FGFR2b+ progenitors.
DISCUSSION
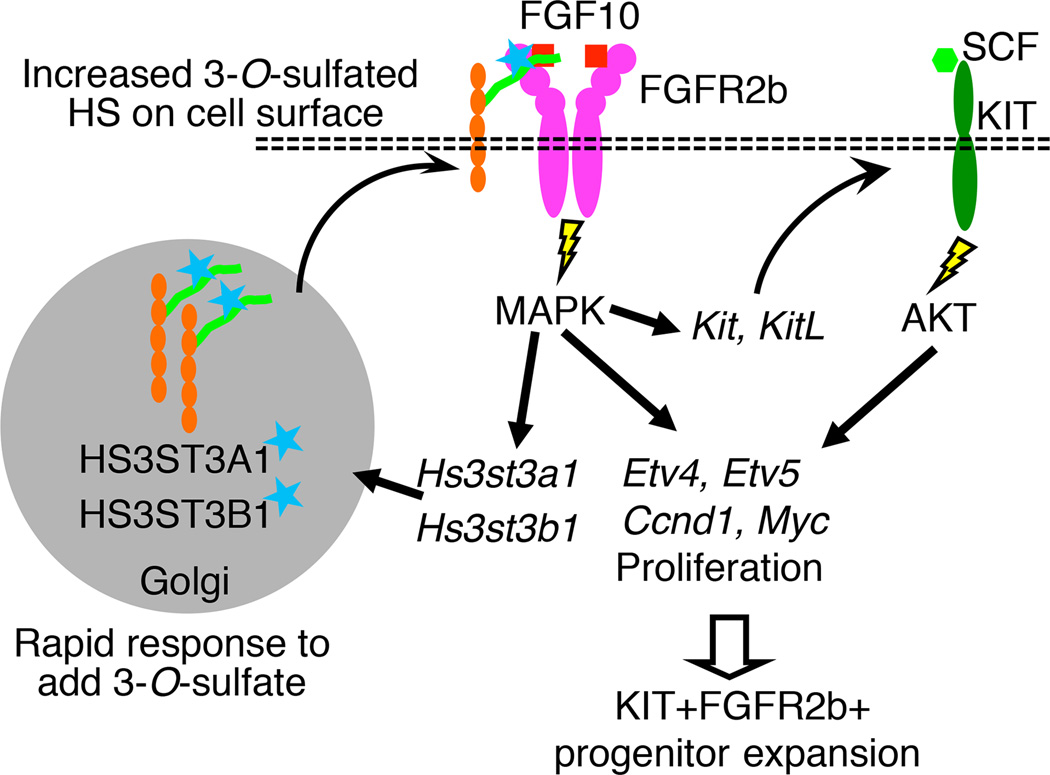
Here we propose a model where 3-O-HS stabilizes FGF10/FGFR2b complex formation to increase FGFR2b-dependent MAPK signaling, gene expression and proliferation (Figure 7). The proliferative response of KIT+FGFR2b+ progenitors is enhanced and maintained via rapid feedback to increase Hs3st expression. Thus, 3-O-HS provides a cellular mechanism to respond to a specific growth factor and amplify that response to expand specific progenitors. The function of specific HS modifications fine-tuning growth factor signaling via regulation of HS biosynthetic enzymes is an emerging paradigm supported by other studies. For example, a 6-O-endosulfatase fine-tunes Hedgehog (Hh) patterning activity of the Drosophila wing disc and this involves regulation of the sulfatase expression pattern by EGFR signaling (Wojcinski et al., 2011). In addition, during lacrimal gland development N-sulfation potentiates FGF10/FGFR2 interactions and Shp2/Erk signaling, which feedback to regulate Ndst expression (Pan et al., 2008).
Figure 7.
Rapidly altering 3-O-sulfation provides a cellular mechanism to modulate the response to FGFR2b signaling and control progenitor expansion. Model showing that 3-O-HS rapidly increases 3-O-sulfotranseferase expression, 3-O-sulfated HS on the epithelium and FGF10/FGFR2b complex formation, MAPK signaling, gene expression of FGFR2b signaling targets, proliferation genes, and Kit and its ligand, which expands the KIT+FGFR2b+ progenitors.
The 3-O-sulfotransferase family creates the least abundant type of sulfate modification, making it difficult to isolate and analyze these rare 3-O-sulfated products. Currently, there is a lack of 3-O-sulfated standards for biochemical analysis, and only recently the enzyme that specifically removes 3-O-sulfation, arylsulfatase G, was identified (Kowalewski et al., 2012). Research has focused on Hs3st1 because it forms the AT-binding HS epitope, but here we used a bioengineered 3-O-HS to investigate Hs3st3 function. Treatment of kidney HS, which has low endogenous 3-O-sulfation, with recombinant 3-O-sulfotransferases enzymes added ~2 extra sulfates/HS chain and allowed us to probe specific functions of 3st3-HS. While HS modified by both Hs3st1 and Hs3st3 have some similar functions, the specific epitopes on these HS chains responsible for specific activity in a particular assay remain to be defined, and epitopes may have some overlapping functions in specific assays, or multiple epitopes may influence a particular function. Our data support the hypothesis that the sulfate pattern, and not just overall charge, is critical for specific HS functions and fine-tuning of growth factor signaling.
The rapid response to 3-O-HS in KIT+FGFR2b+ progenitors may be due to a number of factors. Firstly, we show that 3-O-HS binds FGFR2b, which stabilizes the FGF10/FGFR2b complex, and this may affect the FGF binding affinity. The binding affinity of FGF10 to HS and FGFR2b are crucial for its biological outcome. A mutation in the HS binding site of FGF10, reduces its binding to HS converting FGF10 into a functional mimic of FGF7, in terms of organ morphogenesis and global gene expression. Whereas, a mutation in FGF10 binding site for FGFR2b, reduces the overall growth response (Makarenkova et al., 2009). Secondly, the binding of FGF10 to FGFR2b increases endocytic recycling of FGFR2b, which correlates with higher mitogenic activity, whereas the binding of FGF7 results in receptor ubiquitination and degradation (Belleudi et al., 2007). We speculate that formation of 3-O-HS/FGF10/FGFR2b complexes combined with endocytic FGFR recycling, would increase and maintain a mitotic response in KIT+FGFR2b+ progenitors.
We propose that a similar mechanism involving 3-O-HS may exist in other organs that develop via FGFR2b-dependent branching morphogenesis such as the lung, kidney, mammary gland, pancreas, lacrimal gland and prostate gland (Davies, 2002; Hogan and Kolodziej, 2002; Tsau et al., 2011). Since multiple Hs3sts are expressed in a variety of tissues (Figure S1D), some redundancy or overlap in their function may exist. However, for the lung, we show that 3-O-HS has similar effects on epithelial endbuds (Figure S3), which also express KIT+FGFR2b+ progenitors (Lombaert et al., 2013). Lacrimal gland endbud proliferation is dependent on FGF10 and NDST1 (Pan et al., 2008). However, NDST1 acts early in HS biosynthesis, and the effects of reducing N-sulfation on 3-O-sulfation were not examined. Thus, unique HS-modifications may alter tissue-specific progenitor expansion.
Since Hs3sts modulate growth factor responses, it is not surprising that they influence tumor progression (Bishop et al., 2007). Extensive DNA methylated sites are present in HS3ST1, HS3ST3A1 and HS3ST6 in HEMC chondrosarcoma cells that repress HS3ST expression and enhance the invasive phenotype (Bui et al., 2010). Hypermethylation of Hs3st2 also reduces it's expression and occurs in a wide range of human cancers including breast, colon, lung, and pancreatic cancers (Miyamoto et al., 2003). Thus, tumorogeneisis correlates with reduced HS3ST-enzymes, which may disrupt the specificity and fine-tuning of growth factor responses. It will be interesting to determine whether 3-O-HS can modify growth factor responses of cancer cells.
Apart from expanding progenitors, HS sulfation also influences progenitor fate (Cool and Nurcombe, 2006). Studies of mouse embryonic stem cells (ESCs) with HS biosynthetic enzymes deleted, show there are effects on pluripotency and FGF and BMP-dependent differentiation (Kraushaar et al., 2012; Lanner et al., 2010). Yet, in vitro addition of HS saccharides of specific length and sulfation directed BMP-dependent ESC differentiation and FGF-dependent neuronal cell differentiation (Holley et al., 2011; Pickford et al., 2011). Additionally, an embryonic form of HS, whose HS structure was not identified, enhanced the multipotency of human mesenchymal stem cells (Helledie et al., 2012). Our data shows that 3-O-HS can expand both fetal and adult KIT+FGFR2b+ progenitors in vitro, but it is unclear whether it influences their cell fate or potency. Other HS-dependent growth factor pathways may be involved. Additionally, heparanases or sheddases, which can release 3-O-HS from the KIT+ progenitor surface, may also influence the fate of adjacent progenitors.
An important question remains as to the identity of core proteins that have 3-O-HS added to them. Our FACS data show increased, but not significant, expression of syndecan 4 (Sdc4) in KIT+ progenitors (Figure S1A). Our temporal expression data, however, shows that Hs3st gene expression increases within 1–2 hr, but that the increase in core protein expression occurs later, after 6 hr (data not shown). This suggests the cell could first increase enzyme synthesis to modify the pool of HS being synthesized, but then also upregulate specific core proteins later. Indeed, 3-O-HS production is regulated by limiting levels of HS3ST enzymes that modify only a portion of an excess precursor pool (Shworak et al., 1996; Yabe et al., 2001). Moreover within a clonal cell line, HS structure appears to be equivalent across multiple core proteins (Shworak et al., 1994). It is unclear whether syndecan 4 HS is specifically 3-O-sulfated in response to FGF10, and the dynamics of HSPG core synthesis and coordination with changes in the HS biosynthetic program require further investigation.
In conclusion, we show that the fine-tuning of HS biosynthesis at its terminal step, by adding the least abundant sulfate group, ensures a rapid and specific response to FGF10 signals. Autocrine feedback, from 3-O-HS/FGF10/FGFR2b signaling, upregulates Hs3st3 expression, to increase 3-O-HS and maintain the proliferation of epithelial KIT+FGFR2b+ progenitors. We propose that 3-O-HS may increase the expansion of KIT+FGFR2b+ progenitors from patient biopsies in vitro for use in autologous progenitor transplantation to regenerate organs.
EXPERIMENTAL PROCEDURES
Ex Vivo Organ Culture and Fluorescence-activated cell sorting
Fetal SMG or lung organ cultures and FACS analysis were recently described in (Lombaert et al., 2013). Single cell suspensions of SMGS were incubated with anti-KIT-APC (BD Pharmingen) and anti-ECAD-PE (R&D), and sorted on a BD Aria II.
Labeling of HS
AT-488 was prepared as previously described (Girardin et al., 2005). We used HSV-1gD, which has a truncated ectodomain at residues 285 and binds to receptors in vitro with a 100-fold increased affinity than full-length ectodomains (Krummenacher et al., 1999). For details, see Extended Experimental Procedures.
Preparation of 3-OST-modified HS
The 3-OST-modified HS was prepared by incubating HS (from bovine kidney) and purified 3-OST-1 and 3-OST-3 enzymes as described (Xu et al., 2008). Briefly, 10 µg HS, 100 mM PAPS, 50 mM MES, pH 7.0, 10 mM MnCl2, 5 mM MgCl2, and 1% Triton X-100(v/v) and 4 mg of 3-OST-1 or 3-OST-3 proteins were incubated in 100 µl at 37°C for 1 hr. The 3-OST-modified HS was isolated by a DEAE-Sephacel chromatography. To monitor the extent of 3-O-sulfation, 3-O-[35S] sulfated HS was prepared using [35S] PAPS (specific activity 6500 cpm/nmole, 100 mM). Disaccharide analysis of the modified HS and binding to AT and gD1 were described (Xu et al 2008). Typically, ~two 3-O-sulfo groups per polysaccharide were incorporated.
Quantitative PCR (qPCR)
Real time PCR was performed as previously described (Patel et al., 2008). Briefly, cDNA was generated from DNase-free RNA, amplified and gene expression normalized to the housekeeping gene, Rps29. The reactions were run in triplicate, and the experiment repeated three times.
In Situ Hybridization
DIG-labeled RNA probes were prepared using the appropriate RNA polymerase using the DIG RNA labeling Kit (Roche Applied Science) according to the manufacturer's instructions. In situ hybridization was previously described (Patel et al., 2008).
Pull-down assays
Equal molar concentrations (5 nM) of recombinant FGF10, recombinant FGFR2b and HS were incubated together for pull-down assays as described (Sugaya et al., 2008) with slight modification. For details, see Extended Experimental Procedures.
FGFR2b and FGF10/GAG binding assay
An enzyme-based assay was performed to determine the binding of FGFR2b or FGF10 to 3-O-HS using 96-well Heparin/GAG binding plates (Iduron Ltd), according to the manufacturer’s protocol. FGFR2b and FGF10 binding detected as described in the Extended Experimental Procedures.
RNA interference and heparan sulfate rescue
At least two siRNAs targeting each Hs3st gene were purchased (Qiagen or Dharmacon). The siRNA sequences used were: TCA ACC GCA AGT TCT ACC AGA to target Hs3st3a1, Hs3st3b1 and Hs3st6, referred as Hs3st3a-3b-6 (Dharmacon), CAC CTC CTT GTC AAA TCT ATA to target Hs3st1 and AAT TCT CCG AAC GTG TCA CGT the non-silencing control (both Qiagen). Isolated epithelial rudiments cultured in laminin-111 gel were transfected with a combination of Hs3st1 and Hs3st3a-3b-6 (100 nM each) siRNA in 200 µl of culture medium using oligofectamine (Invitrogen) as previously described (Rebustini et al., 2007). HS or 3-O-HS (0.5 µg/ml or 35 nM) was added following 16 hr of siRNA transfection. Gene knockdown was measured by qPCR analysis after 40 hr culture.
Immunofluorescence analysis and detection of cell proliferation
Whole mount immunofluorescence was performed as previously described (Rebustini et al., 2012). SMG epithelial proliferation was detected using Click-iT EdU Alexa Fluor 488 proliferation Kit (Invitrogen) according to the manufacturer’s protocols. Binding of labeled AT488 or gD1 was performed as described in the Extended Experimental Procedures. Images were obtained using a Zeiss LSM 710 confocal microscope.
BaF3 Proliferation assays
BaF3 cell stably expressing the FGFR splice isoforms (FGFR1b, 2b, 2c and 3c) were kindly provided by Dr. David Ornitz (Washington University) and maintained as previously described (Ornitz et al., 1996). For a detailed protocol, see Extended Experimental Procedures.
Salisphere assay
This assay was performed as previously described (Lombaert et al., 2008) with minor modification to the culture medium. Carbachol (10 nM), Y-27632 rock inhibitor (5 µM) (Calbiochem), FGF10 (50 ng/mL) with or without HS or 3st3-HS (50 ng/mL/3.6 nM) was also supplemented to the culture medium. At day 3, the salispheres were counted and photographed to measure their size. The salispheres were cultured for six days and the either lysed for RNA extraction or stained for KIT and CCND1.
Statistical analysis
Data were log transformed before all statistical analysis. We used GraphPad Prism 6 software (GraphPad Software). The Student’s t test was used to compare two different groups, one-way ANOVA with post hoc Dunnett’s test to compare more than two experimental groups, and multiple unpaired t tests were analyzed by a False Discovery Rate approach where Q, the acceptable rate for false discoveries was set to 5%. Graphs show the mean ± SEM for each group from three or more experiments.
Supplementary Material
HIGHLIGHTS.
Epithelial KIT+ progenitors express enzymes that generate 3-O-sulfated HS
3-O-sulfotransferases are rapidly upregulated by FGF10/FGFR2b signaling
3-O-sulfated HS binds FGFR2b and amplifies FGF10/FGFR2b signaling and proliferation
3-O-sulfated HS expands fetal and adult KIT+FGFR2b+ progenitors
ACKNOWLEDGEMENTS
We would like to thank Dr. S.M. Knox and Dr. I.T. Rebustini for helpful discussions, Drs. G.H. Cohen and R.J. Eisenberg (University of Pennsylvania) for the HSV gD1 protein and Dr. David Ornitz (Washington University) for the BaF3 cell lines. The research was supported by the Intramural Research Program of the National Institute of Dental and Craniofacial Research at the NIH and R01 HL079104 to N.W.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTARY INFORMATION
Supplementary Information includes Extended Experimental Procedures, and three figures.
REFERENCES
- Belleudi F, Leone L, Nobili V, Raffa S, Francescangeli F, Maggio M, Morrone S, Marchese C, Torrisi MR. Keratinocyte growth factor receptor ligands target the receptor to different intracellular pathways. Traffic. 2007;8:1854–1872. doi: 10.1111/j.1600-0854.2007.00651.x. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- Bui C, Ouzzine M, Talhaoui I, Sharp S, Prydz K, Coughtrie MW, Fournel-Gigleux S. Epigenetics: methylation-associated repression of heparan sulfate 3-O-sulfotransferase gene expression contributes to the invasive phenotype of H-EMC-SS chondrosarcoma cells. FASEB J. 2010;24:436–450. doi: 10.1096/fj.09-136291. [DOI] [PubMed] [Google Scholar]
- Cool SM, Nurcombe V. Heparan sulfate regulation of progenitor cell fate. J. Cell Biochem. 2006;99:1040–1051. doi: 10.1002/jcb.20936. [DOI] [PubMed] [Google Scholar]
- Davies JA. Do different branching epithelia use a conserved developmental mechanism? Bioessays. 2002;24:937–948. doi: 10.1002/bies.10161. [DOI] [PubMed] [Google Scholar]
- De Moerlooze L, Spencer-Dene B, Revest JM, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Girardin EP, Hajmohammadi S, Birmele B, Helisch A, Shworak NW, de Agostini AI. Synthesis of anticoagulantly active heparan sulfate proteoglycans by glomerular epithelial cells involves multiple 3-O-sulfotransferase isoforms and a limiting precursor pool. J. Biol. Chem. 2005;280:38059–38070. doi: 10.1074/jbc.M507997200. [DOI] [PubMed] [Google Scholar]
- HajMohammadi S, Enjyoji K, Princivalle M, Christi P, Lech M, Beeler D, Rayburn H, Schwartz JJ, Barzagar S, de Agostini AI, Post M, Rosenberg RD, Shworak NW. Normal levels of anticoagulant heparan sulfate are not essential for normal hemostasis. J. Clin. Invest. 2003;111:989–999. doi: 10.1172/JCI15809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H, Wang F. Visualizing mechanosensory endings of TrkC-expressing neurons in HS3ST-2-hPLAP mice. J. Comp. Neurol. 2008;511:543–556. doi: 10.1002/cne.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BL, Kolodziej PA. Organogenesis: molecular mechanisms of tubulogenesis. Nat. Rev. Genet. 2002;3:513–523. doi: 10.1038/nrg840. [DOI] [PubMed] [Google Scholar]
- Holley RJ, Pickford CE, Rushton G, Lacaud G, Gallagher JT, Kouskoff V, Merry CL. Influencing hematopoietic differentiation of mouse embryonic stem cells using soluble heparin and heparan sulfate saccharides. J. Biol. Chem. 2011;286:6241–6252. doi: 10.1074/jbc.M110.178483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura K, Rhodes JM, Ueda R, McNeely M, Shukla D, Kimata K, Spear PG, Shworak NW, Nakato H. Regulation of Notch signaling by Drosophila heparan sulfate 3-O sulfotransferase. J. Cell Biol. 2004;166:1069–1079. doi: 10.1083/jcb.200403077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalewski B, Lamanna WC, Lawrence R, Damme M, Stroobants S, Padva M, Kalus I, Frese MA, Lubke T, Lullmann-Rauch R, et al. Arylsulfatase G inactivation causes loss of heparan sulfate 3-O-sulfatase activity and mucopolysaccharidosis in mice. PNAS. 2012;109:10310–10315. doi: 10.1073/pnas.1202071109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushaar DC, Rai S, Condac E, Nairn A, Zhang S, Yamaguchi Y, Moremen K, Dalton S, Wang L. Heparan sulfate facilitates FGF and BMP signaling to drive mesoderm differentiation of mouse embryonic stem cells. J. Biol. Chem. 2012;287:22691–22700. doi: 10.1074/jbc.M112.368241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummenacher C, Rux AH, Whitbeck JC, Ponce-de-Leon M, Lou H, Baribaud I, Hou W, Zou C, Geraghty RJ, Spear PG, et al. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J. Virol. 1999;73:8127–8137. doi: 10.1128/jvi.73.10.8127-8137.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanner F, Lee KL, Sohl M, Holmborn K, Yang H, Wilbertz J, Poellinger L, Rossant J, Farnebo F. Heparan sulfation-dependent fibroblast growth factor signaling maintains embryonic stem cells primed for differentiation in a heterogeneous state. Stem Cells. 2010;28:191–200. doi: 10.1002/stem.265. [DOI] [PubMed] [Google Scholar]
- Lawrence R, Yabe T, Hajmohammadi S, Rhodes J, McNeely M, Liu J, Lamperti ED, Toselli PA, Lech M, Spear PG, et al. The principal neuronal gD-type 3-O-sulfotransferases and their products in central and peripheral nervous system tissues. Matrix Biol. 2007;26:442–455. doi: 10.1016/j.matbio.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl U, Backstrom G, Thunberg L, Leder IG. Evidence for a 3-O-sulfated D-glucosamine residue in the antithrombin-binding sequence of heparin. PNAS. 1980;77:6551–6555. doi: 10.1073/pnas.77.11.6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Pedersen LC. Anticoagulant heparan sulfate: structural specificity and biosynthesis. Appl. Microbiol. Biotechnol. 2007;74:263–272. doi: 10.1007/s00253-006-0722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert IM, Abrams SR, Li L, Eswarakumar VP, Sethi AJ, Witt RL, Hoffman MP. Combined Kit and FGFR2b signaling regulates epithelial progenitor expansion during organogenesis. Stem Cell Reports. 2013;1:604–619. doi: 10.1016/j.stemcr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert IM, Brunsting JF, Wierenga PK, Faber H, Stokman MA, Kok T, Visser WH, Kampinga HH, de Haan G, Coppes RP. Rescue of salivary gland function after stem cell transplantation in irradiated glands. PLoS One. 2008;3:e2063. doi: 10.1371/journal.pone.0002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarenkova HP, Hoffman MP, Beenken A, Eliseenkova AV, Meech R, Tsau C, Patel VN, Lang RA, Mohammadi M. Differential interactions of FGFs with heparan sulfate control gradient formation and branching morphogenesis. Sci Signal. 2009;2:ra55. doi: 10.1126/scisignal.2000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Asada K, Fukutomi T, Okochi E, Yagi Y, Hasegawa T, Asahara T, Sugimura T, Ushijima T. Methylation-associated silencing of heparan sulfate D-glucosaminyl 3-O-sulfotransferase-2 (3-OST-2) in human breast, colon, lung and pancreatic cancers. Oncogene. 2003;22:274–280. doi: 10.1038/sj.onc.1206146. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Olsen SK, Ibrahimi OA. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Neugebauer JM, Cadwallader AB, Amack JD, Bisgrove BW, Yost HJ. Differential roles for 3-OSTs in the regulation of cilia length and motility. Development. 2013;140:3892–3902. doi: 10.1242/dev.096388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Pan Y, Carbe C, Powers A, Zhang EE, Esko JD, Grobe K, Feng GS, Zhang X. Bud specific N-sulfation of heparan sulfate regulates Shp2-dependent FGF signaling during lacrimal gland induction. Development. 2008;135:301–310. doi: 10.1242/dev.014829. [DOI] [PubMed] [Google Scholar]
- Patel VN, Hoffman MP. Salivary Gland Development: A Template for Regeneration. Semin. Cell Dev. Biol. 2013 doi: 10.1016/j.semcdb.2013.12.001. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VN, Likar KM, Zisman-Rozen S, Cowherd SN, Lassiter KS, Sher I, Yates EA, Turnbull JE, Ron D, Hoffman MP. Specific Heparan Sulfate Structures Modulate FGF10-mediated Submandibular Gland Epithelial Morphogenesis and Differentiation. J. Biol. Chem. 2008;283:9308–9317. doi: 10.1074/jbc.M709995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VN, Rebustini IT, Hoffman MP. Salivary gland branching morphogenesis. Differentiation. 2006;74:349–364. doi: 10.1111/j.1432-0436.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- Petiot A, Conti FJ, Grose R, Revest JM, Hodivala-Dilke KM, Dickson C. A crucial role for Fgfr2-IIIb signalling in epidermal development and hair follicle patterning. Development. 2003;130:5493–5501. doi: 10.1242/dev.00788. [DOI] [PubMed] [Google Scholar]
- Pickford CE, Holley RJ, Rushton G, Stavridis MP, Ward CM, Merry CL. Specific glycosaminoglycans modulate neural specification of mouse embryonic stem cells. Stem Cells. 2011;29:629–640. doi: 10.1002/stem.610. [DOI] [PubMed] [Google Scholar]
- Rebustini IT, Hayashi T, Reynolds AD, Dillard ML, Carpenter EM, Hoffman MP. miR-200c regulates FGFR-dependent epithelial proliferation via Vldlr during submandibular gland branching morphogenesis. Development. 2012;139:191–202. doi: 10.1242/dev.070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebustini IT, Patel VN, Stewart JS, Layvey A, Georges-Labouesse E, Miner JH, Hoffman MP. Laminin alpha5 is necessary for submandibular gland epithelial morphogenesis and influences FGFR expression through beta1 integrin signaling. Dev. Biol. 2007;308:15–29. doi: 10.1016/j.ydbio.2007.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Hogan BL. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol. 2011;27:493–512. doi: 10.1146/annurev-cellbio-100109-104040. [DOI] [PubMed] [Google Scholar]
- Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- Shworak NW, Fritze LM, Liu J, Butler LD, Rosenberg RD. Cell-free synthesis of anticoagulant heparan sulfate reveals a limiting converting activity that modifies an excess precursor pool. J. Biol. Chem. 1996;271:27063–27071. [PubMed] [Google Scholar]
- Shworak NW, Shirakawa M, Colliec-Jouault S, Liu J, Mulligan RC, Birinyi LK, Rosenberg RD. Pathway-specific regulation of the synthesis of anticoagulantly active heparan sulfate. J. Biol. Chem. 1994;269:24941–24952. [PubMed] [Google Scholar]
- Sugaya N, Habuchi H, Nagai N, Ashikari-Hada S, Kimata K. 6-O-sulfation of heparan sulfate differentially regulates various fibroblast growth factor-dependent signalings in culture. J. Biol. Chem. 2008;283:10366–10376. doi: 10.1074/jbc.M705948200. [DOI] [PubMed] [Google Scholar]
- Tecle E, Diaz-Balzac CA, Bulow HE. Distinct 3-O-sulfated heparan sulfate modification patterns are required for kal-1-dependent neurite branching in a context-dependent manner in Caenorhabditis elegans. G3. 2013;3:541–552. doi: 10.1534/g3.112.005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsau C, Ito M, Gromova A, Hoffman MP, Meech R, Makarenkova HP. Barx2 and Fgf10 regulate ocular glands branching morphogenesis by controlling extracellular matrix remodeling. Development. 2011;138:3307–3317. doi: 10.1242/dev.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcinski A, Nakato H, Soula C, Glise B. DSulfatase-1 fine-tunes Hedgehog patterning activity through a novel regulatory feedback loop. Dev. Biol. 2011;385:168–180. doi: 10.1016/j.ydbio.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Moon AF, Song D, Pedersen LC, Liu J. Engineering sulfotransferases to modify heparan sulfate. Nat. Chem. Biol. 2008;4:200–202. doi: 10.1038/nchembio.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T, Shukla D, Spear PG, Rosenberg RD, Seeberger PH, Shworak NW. Portable sulphotransferase domain determines sequence specificity of heparan sulphate 3-O-sulphotransferases. Biochem J. 2001;359:235–241. doi: 10.1042/0264-6021:3590235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.