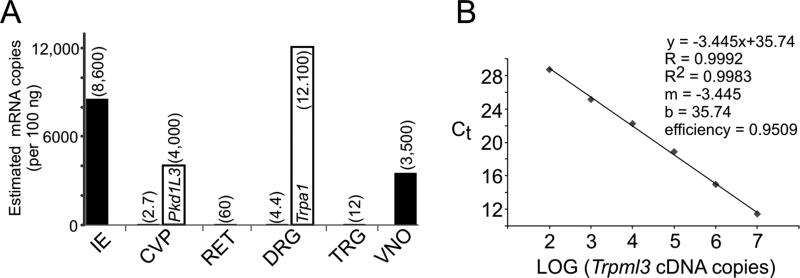
Figure 6.
Levels of Trpml3 mRNA in different sensory organs estimated by RT-qPCR. A,B: One microgram of total RNA from each sensory organ was reverse transcribed, and one-tenth of the resulting cDNA served as template for quantitative PCR. The values per sensory organ are the average of three separate reactions, except for Trpml3 in retina and in circumvallate papillae, for which only one of the three reactions detected any mRNA, so the number given should be considered an upper estimate. Only results from Trpml3+/+ tissues are shown, because no amplification product was obtained from Trpml3−/− tissues, confirming the specificity of the product. 18S rRNA was used as the reference gene, and VNO, which had the highest amounts of 18S rRNA and thus probably the least degraded RNA, was used as the tissue calibrator by means of 2−ΔΔCt. By comparing with qPCR values from known copy numbers (A; over six orders of magnitude) of Trpml3 cDNA template, we estimated the number of mRNAs in each tissue sample (A; indicated in parenthesis over each bar). Sensory organs or tissues examined were inner ear (IE), circumvallate papillae with adjacent tongue tissue (CVP), retina without pigmented epithelium and choroid (RET), L4 dorsal root ganglion (DRG), trigeminal ganglion (TRG), and vomeronasal organ (VNO). Open bar for CVP indicates estimated copy number of Pkd1l3 mRNA, and open bar for DRG indicates estimated copy number of Trpa1 mRNA.