Figure 1.
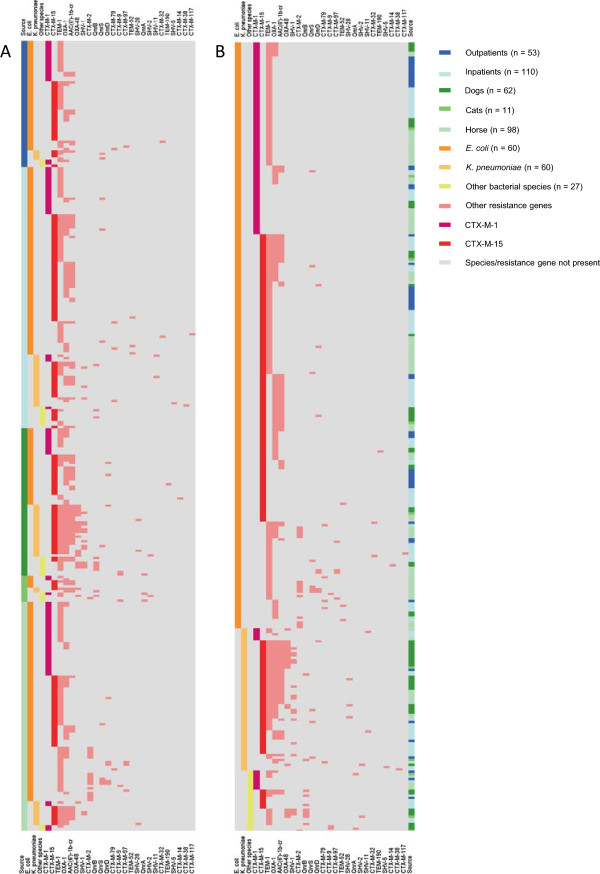
Heat maps generated from identified resistance genes and bacterial species among human and animal isolates. Identified resistance genes and bacterial species are listed according to frequency (except for CTX-M-1 and CTX-M-15) on the right and left side of the figures. Source = origin of isolates (outpatients, inpatients, dogs, cats, horses). Not included are isolates without detectable resistance gene (n = 28). The term “Other species” includes Enterobacter cloacae (n = 12), Klebsiella oxytoca (n = 9), Enterobacter intermedius (n = 2), Enterobacter gergoviae (n = 1), Citrobacter freundii (n = 1) and Proteus mirabilis (n = 1). A is focussing on the origin of isolates whereas the B emphasizes the involved bacterial species.
