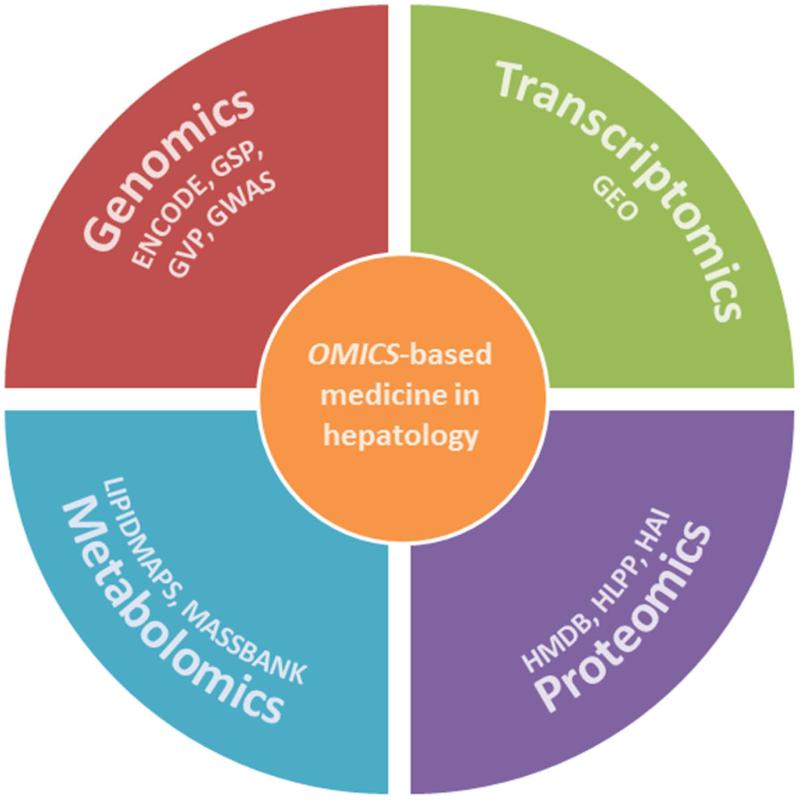
Figure 3. Omics-based medicine.
The ultimate aim of omics-based medicine is to translate human genomics, transcriptomics, proteomics and metabolomics results into clinically useful products. To help researchers achieve this goal several freely accessible initiatives have been established, such as the Genome Sequencing Program (GSP), the Encyclopedia of DNA Elements (ENCODE), the Genetic Variation Program (GVP), or the Genome-Wide Associations Studies (GWAS) of the National Human Genome Research Institute (http://www.genome.gov/). In transcriptomics, the Gene Expression Omnibus (GEO) provides a public repository that archives and freely distributes (http://www.ncbi.nlm.nih.gov/geo/info/overview.html) microarrays and other functional genomics data. In proteomics, the Human Proteome Organization (HUPO, http://www.hupo.org/initiatives/) sponsors several initiatives such as the Human Liver Protein Project (HLPP) or the Human Antibody Initiative (HAI); and in metabolomics the Human Metabolome Database (HMDB, http://www.hmdb.ca/) and other related resources such as KEGG, LipidMaps, and MassBank, contain freely available information about metabolites found in the human body,