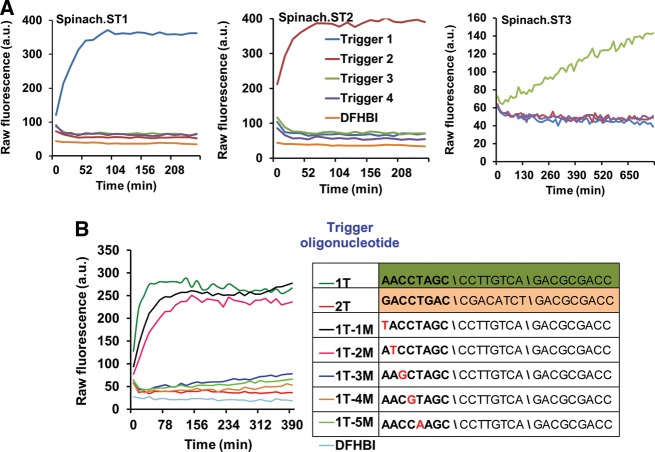
FIGURE 2.
Sequence-dependent activation of Spinach.ST molecular beacons. (A) The molecular beacons Spinach.ST1, Spinach.ST2, and Spinach.ST3 were designed to be specific for three different target nucleic acid sequences. They are activated by only their complementary trigger oligonucleotides Trigger 1, Trigger 2, and Trigger 3, respectively. Spinach.ST RNAs were synthesized by T7 RNA polymerase-mediated transcription of 500 ng of PCR-generated duplex DNA transcription templates. Spinach.ST transcripts were incubated with 0.5 μM trigger DNA oligonucleotides and Spinach.ST activation was measured as fluorescence accumulation over time at 37°C. Raw fluorescence values are shown in arbitrary units (a.u.) and data representative of replicate experiments are depicted. (B) Spinach.ST molecular beacons can detect single-nucleotide mismatches in their target oligonucleotides. Spinach.ST1 transcripts were incubated with 0.7 μM cognate trigger oligonucleotides that were either fully complementary (1T) or contained a single-base mismatch at various positions within the toehold domain 2* (1T-1M to 1T-5M). Trigger sequences with 2*–5*–6* domain organization are depicted in the 5′–3′ direction with mismatches highlighted in red. Spinach.ST1 activation was measured in real-time at 37°C. Incubation with a noncognate trigger oligonucleotide (2T) served as the negative control for Spinach.ST1 activation. Raw fluorescence values are shown in arbitrary units (a.u.) and data representative of replicate experiments are depicted.