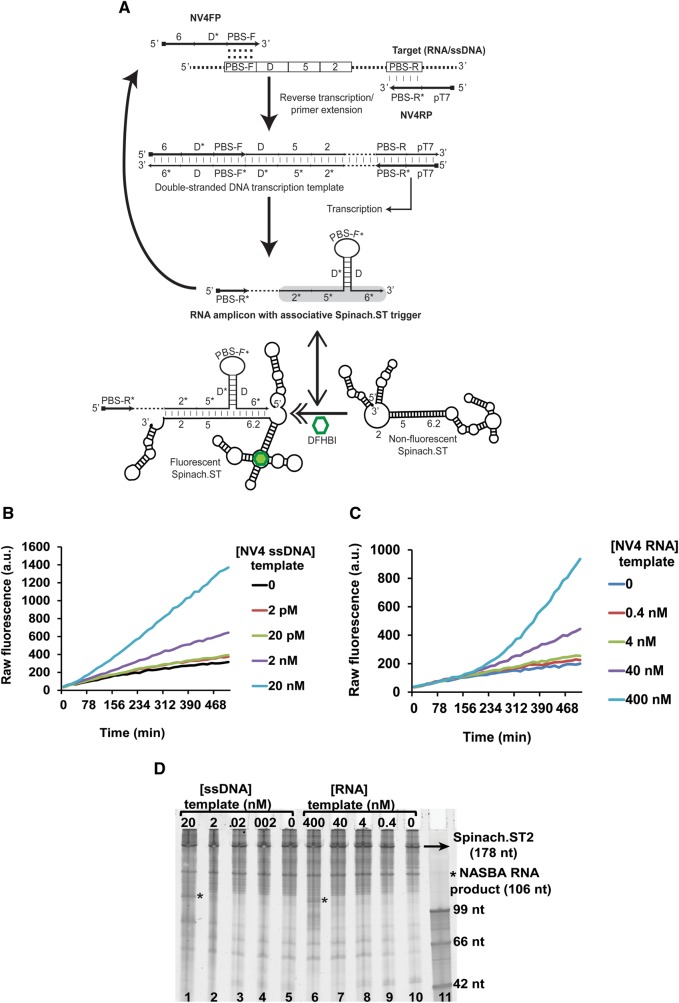
FIGURE 3.
Application of Spinach.ST to the detection of NASBA amplicons. (A) Schematic of primer design for real-time, sequence-specific NASBA detection using Spinach.ST2. Sequence blocks are depicted as numbered or lettered domains (* indicates complementarity). NV4FP and NV4RP are the forward and reverse primers used for NASBA. PBS-F and PBS-R* domains hybridize with the target nucleic acid and initiate polymerization. Hybridization is depicted using vertical slashes, while the potential hybridization of the PBS-F domain within the NV4FP primer to a complementary region is indicated by colons. Dashed regions in the target, double-stranded DNA transcription template, and RNA transcript denote target sequences not involved in priming or in Spinach.ST activation. The Spinach.ST trigger in the NASBA RNA amplicons is highlighted in gray. Spinach.ST structures were generated using NUPACK. (B,C) Sequence-specific fluorescent detection of single-stranded DNA (B) or RNA (C) templates by real-time Spinach.ST-NASBA. Varying concentrations of NV4 template DNA or RNA were amplified at 37°C by NASBA using T7 RNA polymerase and MMLV RT in the presence of cotranscribed Spinach.ST2 and 40 µM DFHBI. Raw fluorescence values are shown in arbitrary units (a.u.) and data representative of replicate experiments are depicted. (D) Denaturing 10% polyacrylamide gel analysis of NASBA amplification reactions of single-stranded DNA (lanes 1–5) or RNA (lanes 6–10) templates; cotranscription of Spinach.ST2 is also shown. Template concentrations are in nanomolar (nM) amounts. Two microliters of each NASBA reaction were analyzed. Single-stranded DNA oligonucleotides were used as size markers (lane 11).