FIGURE 4.
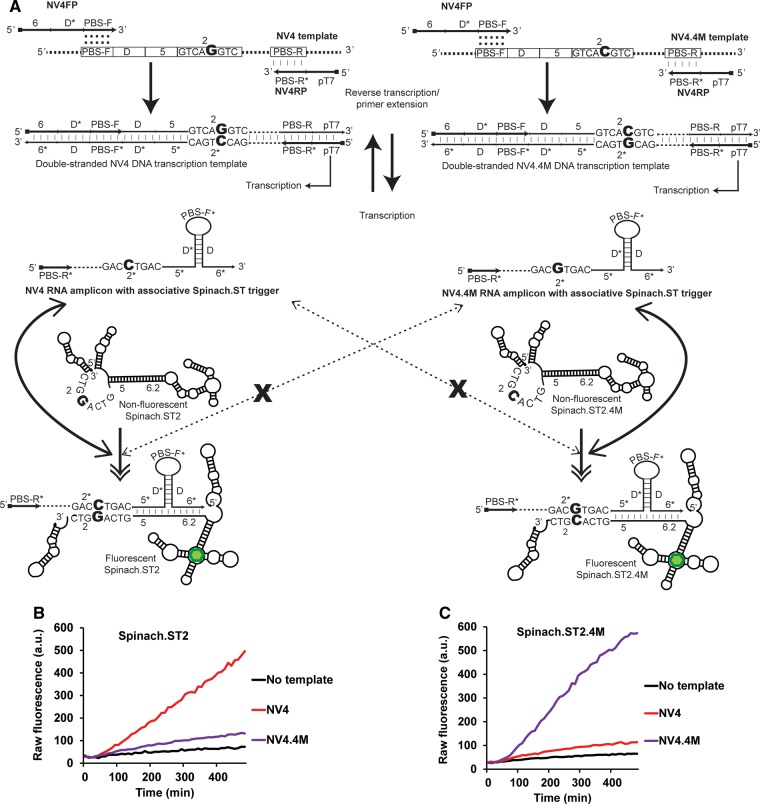
Discrimination of alleles in real-time by NASBA and sequence-modulated Spinach. (A) Schematic depicting the target alleles NV4 and the SNP-containing NV4.4M. Only the domain 2 and 2* sequences are shown for clarity. The single-nucleotide difference between the two alleles (highlighted in a large bold font) is located within the target region termed domain 2. Both NV4 and NV4.4M alleles were amplified using the common primers NV4FP and NV4RP. NASBA-generated NV4 and NV4.4M RNA amplicons form associative Spinach.ST triggers that have a single base difference between them (highlighted in bold font) in their 2* domains. The sequence-modulated molecular beacons Spinach.ST2 and the SNP-complementary Spinach.ST2.4M have corresponding single-nucleotide differences (highlighted in a large bold font) in their toehold domain 2. RNA amplicons bearing fully complementary associative triggers should activate Spinach.ST molecular beacons by facilitating toehold-mediated strand displacement. Activated Spinach.ST molecules increase the fluorescence of DFHBI (depicted as a green hexagon). Activation of Spinach.ST molecules by RNA amplicons bearing a single-nucleotide mismatch in their toehold domain should be significantly impaired (denoted as dashed double-tipped arrows interrupted with an X). (B,C) Detection of NASBA amplicons by allele-specific, sequence-modulated Spinach.ST. NASBA reactions were used to amplify either no template (black line) or 20 nM of ssDNA targets NV4 (red line) or NV4.4M (purple line) in the presence of 40 µM DFHBI and 138 ng of dsDNA transcription templates for either Spinach.ST2 (B) or Spinach.ST2.4M (C). Raw fluorescence values are shown in arbitrary units (a.u.) and data representative of replicate experiments are depicted.