Figure 4.
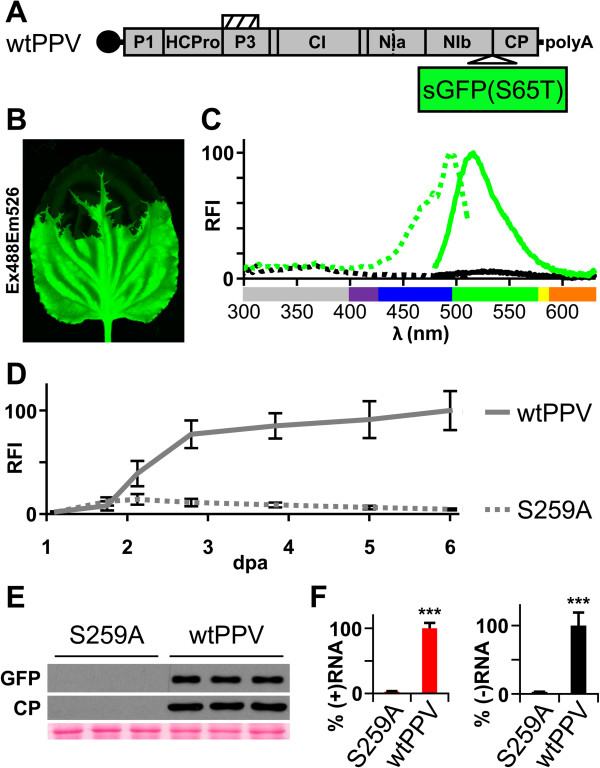
Monitoring of GFP-tagged virus amplification dynamics by fluorescence spectroscopy. GFP-tagged viral cDNA clones pSN-PPV (wtPPV, wild-type PPV) and pSN-PPV P1-S (S259A, in which P1 protease catalytic amino acid S259 was replaced by alanine) were delivered to plants by agro-infiltration. (A) Diagram of wild-type PPV (wtPPV) genome originated following pSN-PPV agro-infiltration. Hatched box indicates P3N-PIPO protein. The reporter sGFP(S65T) gene is inserted between NIb and CP coding sequences. (B) N. benthamiana plants were challenged with pSN-PPV, and fluorescence of systemically infected leaves was detected by laser scanning (10 dpa; green). (C) Excitation (dotted lines) and emission spectra (solid lines) of sGFP(S65T) were measured from pSN-PPV agro-inoculated leaf discs (green); leaves infiltrated with an Agrobacterium culture without expression vectors were used as control (black). Relative fluorescence intensity (RFI) was plotted using sGFP(S65T) peaks equal to 100. UV wavelengths are in gray, visible spectrum colors were assigned as described [39]. (D) GFP fluorescent intensity (RFI) from infiltrated leaves was quantified in a 96-well plate reader and plotted using average wtPPV value at 6 dpa equal to 100. Line graph shows mean ± SD (n = 16 samples/condition, from two independent Agrobacterium cultures). (E) Amount of GFP protein and PPV CP in infiltrated leaves at 6 dpa was assessed by immunoblot analysis. Each lane represents a pool of 3 or 4 leaf samples infiltrated with two independent Agrobacterium cultures. Ponceau red-stained blot is shown as loading control. (F) Amount of viral (+)RNA and (−)RNA from inoculated leaves at 6 dpa was quantified by RT-qPCR and plotted using average wtPPV value equal to 100. Bar graph shows mean ± SD (n = 4 biological replicates, from two independent Agrobacterium cultures); ***p < 0.001, Student’s t-test.
