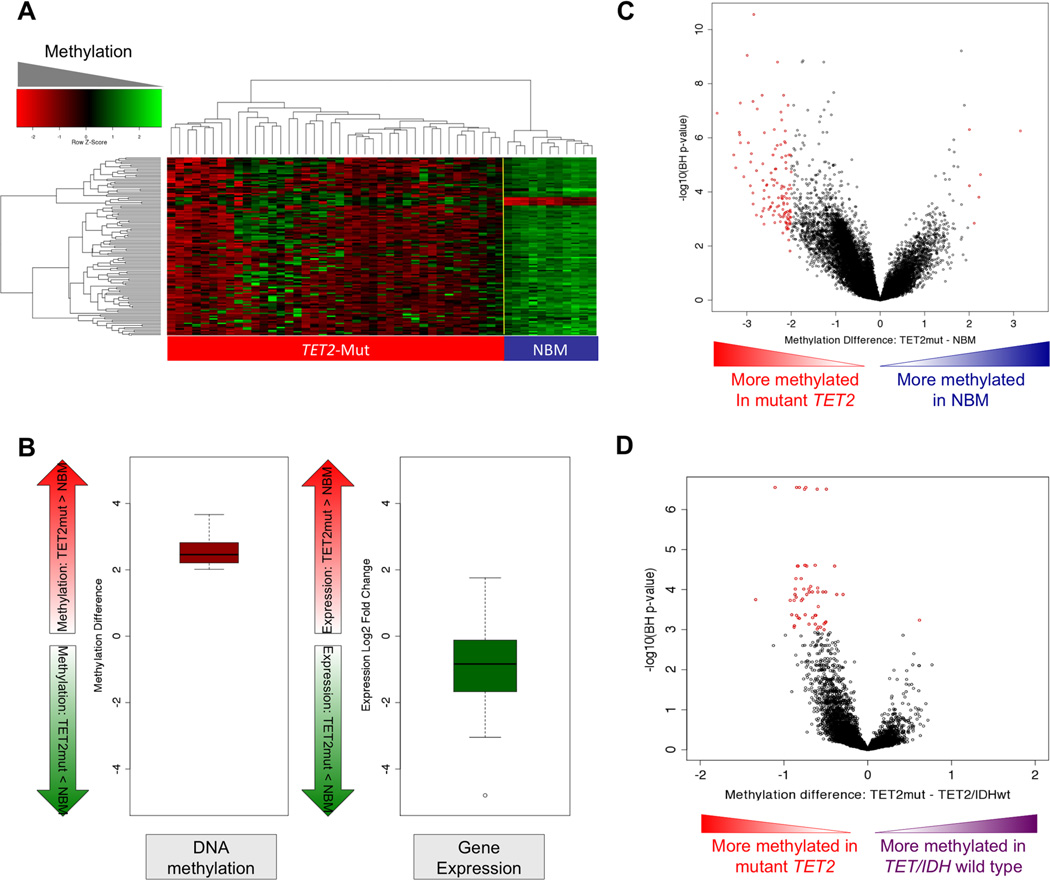
Figure 6. TET2-mutant AML is associated with a hypermethylation phenotype.
(A) Heatmap representation of a two-dimensional hierarchical clustering of genes identified as differentially methylated between TET2 mutant primary AML cases (Mut; red bar) and normal CD34+ bone marrow cells (NBM; blue bar). Each row represents a probe set and each column represents a patient. (B) Boxplot illustrating average methylation difference between TET2 mutant AMLs vs. normal CD34+ cells (left) and average gene expression difference between TET2 mutant AMLs vs. normal CD34+ cells (right) of genes aberrantly methylated in TET2 mutant AMLs. (C) Dot plot of methylation difference between TET2-mutant AMLs and normal CD34+ bone marrow cells (biological significance) vs. statistical significance (-log10 (BH p value)). Red points indicate probe sets identified as differentially methylated between the two groups. (D) Dot plot of methylation difference between TET2-mutant AMLs and TET2 and IDH1/2- wild-type AMLs (biological significance) vs. statistical significance (-log10 (T+BH)). Red points indicate probe sets statistically significant between the two groups. (See also table S3A–C).