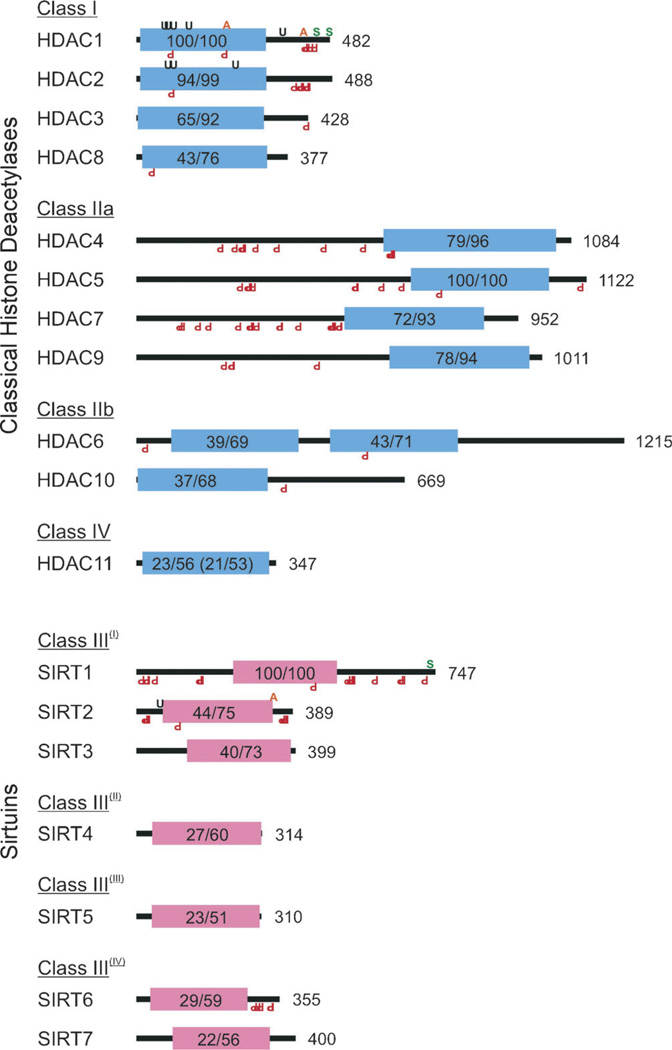
Figure 1.
Schematic representations of human histone deacetylases (HDACs) with the locations of their conserved deacetylase domains; their respective superfamilies are indicated through the color of their deacetylase domains. Both classical HDACs and sirtuins are organized by homology class. Shown are Zn2+ (blue) and NAD+-dependent (red) deacetylase domains, their percentage identity and similarity to a class archetype (e.g., HDAC1, HDAC5, SIRT1), and total protein length in amino acids. HDAC11 has homology to both HDAC1 and HDAC5; the latter values are shown in parentheses. Also indicated are the locations of validated KDAC post-translation modifications including acetylation (orange A, top), SUMOylation (green S), ubiquitination (black U), and phosphorylation (red inverted P, bottom).