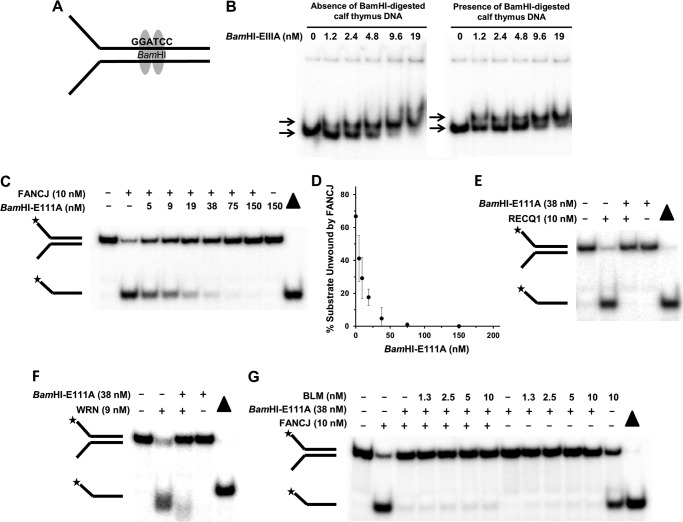
FIGURE 1.
BamHI-E111A bound to forked duplex DNA substrate blocks DNA unwinding by human DNA repair helicases. A, schematic representation of BamHI dimer bound to the palindromic recognition sequence harbored within the duplex region of forked DNA substrate. B, representative gel images from EMSA of reaction mixtures containing the indicated concentrations of BamHI-E111A protein and 0.5 nm radiolabeled forked duplex DNA in the absence or presence of 5 μg/ml BamHI-digested calf thymus DNA. The migration of the protein-free radiolabeled forked duplex and BamHI-E111A-bound forked duplex are indicated by the top and bottom arrows, respectively. C, BamHI-E111A inhibits FANCJ helicase activity on forked duplex DNA substrate in a protein concentration-dependent manner. Representative gel of resolved proteinase K-digested products is shown. D, quantitative analysis of BamHI-E111A inhibition of FANCJ helicase activity. E and F, BamHI-E111A (38 nm) inhibits RECQ1 (E) or WRN (F) helicase activity on forked duplex substrate. G, combination of BLM and FANCJ helicases fail to displace BamHI-E111A from the DNA substrate. Proteinase K-digested products from helicase reaction mixtures were resolved by electrophoresis on nondenaturing 12% polyacrylamide gels. Filled triangle, heat-denatured DNA substrate control. Representative gel images from at least three independent experiments are shown. Star denotes 5′-32P end label.