FIGURE 3.
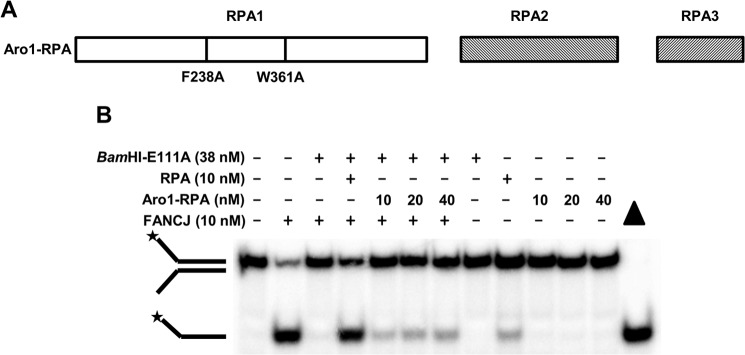
Amino acid substitutions that inactivate DNA binding by RPA negatively affect its ability to stimulate FANCJ disruption of BamHI-E111A protein-DNA substrate interaction. A, schematic representation of Aro1-RPA characterized by two amino acid substitutions in the RPA1 (RPA 70 kDa) subunit of the RPA heterotrimer consisting of RPA1, RPA2, and RPA3. The Aro1-RPA mutant harboring the two amino acid substitutions F238A and W361A was previously shown to inactivate DNA binding by RPA (27). B, reaction mixtures containing BamHI-E111A-bound forked duplex DNA substrate, FANCJ, and either wild-type RPA (RPA) or mutant Aro1-RPA were incubated and analyzed as described under “Experimental Procedures.” Representative gel image showing proteinase K-digested products from at least three independent experiments is shown. Star denotes 5′-32P end label.
