Background: The mechanism of prosaposin processing to generate saposins A-D is unknown.
Results: Epidermis-specific mesotrypsin and caspase-14 generated mature saposins from prosaposin. Deficiency of prosaposin or saposin A resulted in loss of intercellular lipid components necessary for maintenance of skin permeability barrier properties.
Conclusion: Mesotrypsin and caspase-14 are involved in prosaposin processing.
Significance: Saposin generation in epidermis is regulated in a differentiation-associated manner.
Keywords: Caspase; Cell Differentiation; Kallikrein; Keratinocyte; Protein Processing; Bar; Caspase,
Abstract
A proteomics-based search for molecules interacting with caspase-14 identified prosaposin and epidermal mesotrypsin as candidates. Prosaposin is a precursor of four sphingolipid activator proteins (saposins A–D) that are essential for lysosomal hydrolysis of sphingolipids. Thus, we hypothesized that caspase-14 and mesotrypsin participate in processing of prosaposin. Because we identified a saposin A sequence as an interactor with these proteases, we prepared a specific antibody to saposin A and focused on saposin A-related physiological reactions. We found that mesotrypsin generated saposins A–D from prosaposin, and mature caspase-14 contributed to this process by activating mesotrypsinogen to mesotrypsin. Knockdown of these proteases markedly down-regulated saposin A synthesis in skin equivalent models. Saposin A was localized in granular cells, whereas prosaposin was present in the upper layer of human epidermis. The proximity ligation assay confirmed interaction between prosaposin, caspase-14, and mesotrypsin in the granular layer. Oil Red staining showed that the lipid envelope was significantly reduced in the cornified layer of skin from saposin A-deficient mice. Ultrastructural studies revealed severely disorganized cornified layer structure in both prosaposin- and saposin A-deficient mice. Overall, our results indicate that epidermal mesotrypsin and caspase-14 work cooperatively in prosaposin processing. We propose that they thereby contribute to permeability barrier formation in vivo.
Introduction
Caspase-14 is a member of the papain-like cysteine proteases and is exclusively expressed in epidermis and its appendages (1, 2). Recent investigations showed that null mutation of caspase-14 caused a defect of filaggrin degradation and increased trans-epidermal water loss, indicating involvement of caspase-14 in epidermal barrier function (3). However, its physiological functions are not yet fully understood.
Previously we purified an active form of caspase-14 from human corneocyte extract and determined its primary structure (4). Sequence analysis demonstrated that activation of caspase-14 would be accomplished by cleavage at Asp-146. We prepared a cleavage site-directed antibody, h14D146, which recognizes only active caspase-14 (p17/p11). Down-regulation of caspase-14, especially its active form, was evident in skin of patients with atopic dermatitis, which is characterized by impairment of the epidermal barrier function. Activation of caspase-14 occurs at the onset of corneocyte formation (5). We have shown that caspase-14 is completely different from other caspases, being regulated by kallikrein-related peptidase-7 (KLK7) and an intermediate form of caspase-14 (6). KLK7 cleaves procaspase-14 at Tyr-178, transiently generating an intermediate form (p20/p8). This form in turn has the ability to generate the mature form, p17/p11, from procaspase-14. It is not yet clear whether the intermediate form possesses other physiological functions in addition to caspase-14 processing activity.
We have also cloned a novel splicing variant of the PRSS3 gene product, mesotrypsinogen, from a keratinocyte cDNA library (7). We named this trypsinogen 5, because we found that a low level of brain-type trypsinogen 4 is expressed in human keratinocytes. These trypsinogens differ from trypsinogen 3 (mesotrypsinogen) in the N-terminal propeptide. These variants all possess an enterokinase recognition sequence, DDDDKI, and after activation, identical mesotrypsin would be produced. We also observed that expression of enterokinase was confined to the granular layer of human epidermis. Localization of mesotrypsin in the upper epidermis would be consistent with a physiological role in keratinocyte terminal differentiation, which is involved in barrier formation.
The epidermal barrier consists of various components, including natural moisturizing factor (8), extracellular lipid membranes (9), cornified envelope (10), and the keratin-filaggrin complex (11), which serves to support and maintain a flexible yet sturdy architecture. Dramatic changes occur at the transition phase from granular cells to cornified cells. Cellular organelles are digested and the cytoplasm becomes filled with keratin fibers embedded in filaggrin to form macrofibrils. Caspase-14 and mesotrypsin are activated during this transition phase.
Extracellular lipid membrane plays an essential role in permeability barrier formation (9). Lysosomal degradation of various glycosphingolipids by exohydrolases is a critical step for maturation of the lipid membrane, which consists of ceramides, free cholesterol, cholesterol sulfate, free fatty acids, and other molecules (12). During keratinocyte terminal differentiation, glycosphingolipids are synthesized and stored in lamellar bodies. They are processed by a set of lipid hydrolases into more hydrophobic moieties to form extracellular lipid membranes. Interestingly, these hydrolases require cofactors known as saposins (13–15). Saposins are sphingolipid activator proteins that are derived from a single precursor, prosaposin. Discovery of inherited diseases associated with saposin deficiency clearly revealed the essential role of these molecules in sphingolipid metabolism (16, 17). Although each of the saposins is generated by limited proteolysis of prosaposin, the processing enzyme(s) in the epidermis have not yet been identified.
We have been searching for interacting molecules as well as substrates for caspase-14 using liquid chromatography coupled with electrospray tandem mass spectrometry (LC/MS/MS), which is a highly sensitive and powerful method. In the present LC/MS/MS study, we identified caspase-14 and mesotrypsin as interactors with prosaposin. Our results indicate that these proteases participate in the processing of prosaposin to generate mature saposins. We propose that caspase-14 and mesotrypsin are involved in permeability barrier formation in vivo.
EXPERIMENTAL PROCEDURES
Materials
Anti-prosaposin antibody was obtained from Thermo Scientific (Rockford, IL) and glutathione transferase (GST)-prosaposin recombinant protein was purchased from Abnova (Taipei, Taiwan). Anti-trypsin antibody was obtained from Athens Research and Technology, Inc. (Georgia, GA). Recombinant human KLK5,2 KLK7, and recombinant human trypsin 3/PRSS3 (mesotrypsin) were purchased from R&D Systems (Minneapolis, MN). Control siRNA and siRNAs to caspase-14 and mesotrypsin were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies to saposin B, saposin C, and saposin D were from Santa Cruz. Anti-mouse filaggrin IgG was obtained from Covance (Berkley, CA).
Cell Culture and Tissue Specimens
Human keratinocytes derived from normal foreskin were cultured in Humedia KG2 (Kurabo, Osaka, Japan). Human skin specimens were obtained with informed consent from patients undergoing plastic surgery. This study was approved by the Institutional Review Board of Tokyo Medical University and also by the Shiseido Committee on Human Ethics.
Preparation of Anti-saposin A Antibody
Anti-saposin A antibody (anti-SapA Ab) was generated in rabbits using the saposin A peptide, CSYLPVILDIIKGEM, as an antigen. Anti-SapA Ab was purified with antigen peptide affinity chromatography.
Preparation of Procaspase-14, Caspase-14 Intermediate, Active Caspase-14, and Saposin A
cDNA of procaspase-14 was PCR cloned and inserted into pGEX-6P1 vector at the BamHI/XhoI site. GST fusion proteins were purified by means of glutathione-agarose (Qiagen) affinity chromatography. Constitutively active caspase-14 (revC14) and the intermediate form of caspase-14 (revC14-Y178) were constructed using pET15b vector (Novagen) as reported earlier (6). These recombinant proteins were isolated with nickel-agarose and further purified with MonoQ chromatography. revC14 and revC14-Y178 were fully active as measured with synthetic substrates. In addition, GST fusion revC14 was also prepared. GST fusion revC14 and procaspase-14 were used for affinity isolation of caspase-14 interacting proteins. Recombinant saposin A was prepared as follows. Saposin A cDNA was PCR cloned and inserted into pQE30 vector at KpnI/SalI site. Primers used were: forward, CGGGTACCCTTCCCTGCGACATATGCAAAGAC, and reverse, CGCAGTCGACCTACTTCTGGAGAGACTCGCAGAGG. Recombinant saposin A was purified with nickel-agarose chromatography.
Preparation of Tissue and Cell Extracts
Keratinocytes were collected at 80% confluence (growth phase), 100% confluence, 2 days after confluence (120%), 2 days after confluence followed by 1.2 mm calcium treatment (differentiated phase), and after air exposure. Air exposure was done as follows. Keratinocytes were cultured until they were fully confluent. After removal of the medium, keratinocytes were exposed to air for 15 min in a plate with a lid and then incubated for a further 2 days in medium containing 1.2 mm CaCl2. Cell pellets were homogenized using a sintered glass homogenizer (Wheaton Science Products, Millville, NJ) with extraction buffer (10 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1% Nonidet P-40, 5 mm EDTA) and subjected to ultrasonication three times for 10 s. Supernatant was obtained after centrifugation. Extracts from a skin equivalent model (Matrex, Toyobo Co., Osaka, Japan) were prepared similarly. Cornified cell extracts were prepared from scraped heels of healthy individuals as reported previously (4). Each extract was reacted with GST fusion procaspase-14 or revC14 coupled with GST-agarose. After extensive washing, bound proteins were eluted with 100 mm glutathione, dialyzed against H2O, and lyophilized for trypsin digestion.
Processing of Saposin A from Prosaposin by Epidermal Proteases
The effect of epidermal proteases on saposin generation was investigated using recombinant GST-prosaposin. GST-prosaposin was incubated with revC14, revC14-Y178, and mesotrypsin under various conditions. We also used KLK5 and KLK7 as representative trypsin-like and chymotrypsin-like serine proteases, respectively, because they are also expressed in the upper layer of human epidermis (18). Briefly, assay mixtures (40 μl) containing 6 μl of prosaposin (0.15 mg/ml) and 2 μl of each protease (2.5 μg/ml) in appropriate assay buffer were incubated for 30–60 min. Assay buffers used were: 0.1 m HEPES (pH 7.5) + 0.06 m NaCl + 0.01% CHAPS + 5 mm DTT + 1.3 m sodium citrate for revC14, 0.1 m Tris-HCl (pH 8.0) + 0.1% Tween 20 + 5 mm NaClO4 for revC14 and revC14-Y178, and 0.1 m Tris-HCl (pH 8.0) + 1 mm CaCl2 + 0.1% Tween 20 for mesotrypsin, KLK5, and KLK7. After the reaction, an equal amount of 2× SDS sample buffer was added and the mixture heated to 95 °C for 5 min. Degradation products were verified by Western blotting with anti-saposin antibodies. In some experiments, cell extracts prepared from differentiated keratinocytes were used as a source of prosaposin instead of GST-prosaposin.
Activation of Mesotrypsinogen by Caspase-14
Mesotrypsinogen (0.5 μg/ml, 2.5 μl) was preincubated with revC14 (0.875, 1.75, and 3.5 μg/ml, 5 μl) in 20 μl of caspase-14 assay buffer for 30 min at 37 °C. For comparison, enterokinase (0.5 μg/ml, 5 μl) was used similarly. Ten-μl aliquots of the reaction mixtures were removed, mixed with 80 μl of serine protease assay buffer (0.1 m Tris-HCl (pH 8), 0.01% Tween 20), and further incubated for 15 min at 37 °C. After addition of 10 μl of 100 μm Boc-Gln-Ala-Arg-methylcoumarin amide (Peptide Institute, Osaka, Japan), the mixture was incubated for 15 min at 37 °C. The activity of mesotrypsin was measured using an ARVOTM X3 (Perkin Elmer Life Sciences) with excitation at 355 nm and emission at 460 nm. The means of duplicate assays were expressed as relative intensity.
Construction of Skin Equivalent Model
Matrex (Toyobo, Osaka, Japan) was used as a dermal component. Each siRNA was introduced into keratinocytes using RNAi Max (Invitrogen) according to the manufacturer's instructions. After treatment with each siRNA, keratinocytes (40 × 104 cells/200 μl medium) were plated on each well and cultured for 2 days. Medium was removed from the upper well and the skin equivalent model was developed by an additional culture for 10 days in medium consisting of a 1:1 mixture of Humedia KG2 and DMEM + 10% fetal bovine serum.
Quantitative Real-time RT-PCR Analysis
Transcription levels of prosaposin were analyzed by quantitative real-time PCR. Total RNA was extracted from cultured cells with ISOGEN (Nippon Gene, Tokyo, Japan) according to the manufacturer's instructions. The cDNA was reverse-transcribed with SuperScriptTM II (Invitrogen). Real-time RT-PCR was performed on a LightCycler rapid thermal cycler system using a LightCycler 480 SYBR Green I Master (Roche Diagnostics) according to the manufacturer's instructions. Primers used were as follows: forward, GACAATGACATCATGCTGATCAAACTCTC, and reverse, CCTCAAGGAAGCCCACACAGAAC. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a reference gene. Amounts of mRNA were normalized to those of GAPDH and finally presented as ratios to those of an untreated control.
Western Blot Analysis
Extracts from keratinocytes, skin equivalent model, and corneocytes scraped from heels of healthy individuals were separated by SDS-PAGE on 5–20 or 15% gradient gels (e-PAGE E-T520L or SPU-15S, ATTO Co., Tokyo, Japan). After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon-P, Millipore, Bedford, MA) and incubated with the appropriate antibodies. Peroxidase-labeled anti-mouse IgG or anti-rabbit IgG (GE Healthcare) was used as a secondary antibody, and immunoreactive proteins were visualized by chemiluminescence using ECL Plus (GE Healthcare).
Tryptic Digestion of Protein Mixture
The dialyzed protein mixtures were independently suspended in 20 μl of 0.1% RapiGest (Waters Co., Milford, MA) in 50 mm ammonium bicarbonate. After vortexing, the solutions were reduced, alkylated, and digested with l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin as reported previously (19). After the digestion, trifluoroacetic acid was added to the mixture at 0.5% final concentration to halt the reaction and inactivate the RapiGest surfactant.
Identification of Proteins Using Liquid Chromatography Coupled with Electrospray Tandem Mass Spectrometry (LC/MS/MS) Analysis
Digested protein mixtures were subjected to LC/MS/MS-based protein identification analysis as described previously (19, 20). Briefly, the mixtures were loaded by an autosampler (SI-2 semi-micro HPLC system, Shiseido Co., Ltd., Tokyo, Japan) onto a fused silica trapping column (100 μm inner diameter × 1 cm, JupiterProteo C14, 10 μm, Phenomenex, Torrance, CA). The trapping column was desalted with a gradient starting buffer (0.1% formic acid, 5% acetonitrile/purified water) for ∼30 min, then directly connected to a fused silica analytical capillary column (100 μm inner diameter × 12 cm, JupiterProteo C14, 4 μm, Phenomenex) by changing the position of a two-way switching valve. Peptides in the digested mixtures were separated with a 90-min organic gradient (5–75% acetonitrile). The column flow-rate was set to 300-400 nl/min by adjusting the length of a split-resistant capillary (50 μm inner diameter × 50–200 mm). Peptides eluted from the column were directly electrosprayed into a hybrid mass spectrometer (LTQ-Orbitrap, Thermo Fisher Scientific, Waltham, MA). The mass spectrometer was operated in an automatic parallel data-dependent MS/MS mode. After one accurate-mass survey scan by the Orbitrap at 60,000 mass resolution, MS/MS spectra for the largest 5 precursor ions in the survey scan were automatically acquired by the LTQ under the control of the Xcalibur data system (Thermo Fisher Scientific). Collected MS/MS spectra were searched to identify proteins with the SEQUEST algorithm running on Bioworks software (Thermo Fisher Scientific). A non-redundant human protein database (NCBI, 2010) was used for the identification of proteins. Stringent search criteria were used to minimize false identification of proteins (Sf score >0.85, peptide probability >0.001, number of top matches: >1, precursor mass tolerance: <10 ppm, minimum number of peptides to identify proteins: 1, enzyme specificity: half-tryptic or fully tryptic peptides only).
Immunohistochemical Procedures
Dual stainings were carried out using anti-mesotrypsin, h14D146, anti-prosaposin, and anti-SapA primary antibodies. Alexa Fluor 555 or 488 (Molecular Probes Inc., Eugene, OR) was used for fluorescence detection. To visualize nuclei, sections were immersed in 10 ng/ml of DAPI (4′,6′-diamidino-2-phenylindole; Molecular Probes) for 5 min and washed 3 times with PBS.
Proximity Ligation Assay (PLA)
We employed the PLA method to investigate association of interacting molecules in vivo. If two molecules are present in close proximity or are associated, antibodies to these molecules can also be located very close to each other. In this case, secondary antibodies labeled with short complementary DNA chains can be ligated together and their fluorescence signals combined and amplified (as in a PCR). This makes it possible to evaluate the possible association of two molecules in human skin sections.
The PLA reaction was performed using normal rabbit IgG/anti-Prosap Ab, anti-mesotrypsin Ab/h14D146, anti-Prosap Ab/anti-mesotrypsin Ab, and anti-Prosap Ab/h14D146, according to the manufacturer's instructions. The combination of normal rabbit IgG and anti-Prosap Ab served as a negative control.
Oil Red Staining
Frozen sections of skin from prosaposin-deficient and saposin A-deficient mice were incubated with distilled water for 30 s, followed by a 1-min incubation in 60% isopropyl alcohol, and then transferred to Oil Red-O (0.3% solution in isopropyl alcohol) for 15 min at 37 °C. Sections were incubated for 1 min in 60% isopropyl alcohol, lightly stained with hematoxylin, and mounted with glycerin jelly mounting medium (Mount Quick, Daido Sangyo Co., Kanagawa, Japan).
Electron Microscopy
Skin samples were fixed with 4% glutaraldehyde and postfixed with 2% osmium tetroxide (Nacalai Tesque Inc., Kyoto, Japan) for 1 h at 4 °C, followed by dehydration through a graded series of ethanol. They were embedded in Epon 812. Ultrathin sections were cut using an ultramicrotome (Reichert Ultracut S, Reichert, Vienna, Austria), stained with saturated uranyl acetate and lead citrate, and observed under a transmission electron microscope (Hitachi H-7100, Hitachi Co. Ltd., Tokyo, Japan).
Prosaposin-deficient Mouse and Saposin A-deficient Mouse
Prosaposin- and saposin A-deficient mice were generated by means of gene targeting technology as described before (21, 22). The mice were anesthetized with ether and skin samples were obtained from 1-month-old prosaposin-deficient mouse and a 6-month-old saposin A-deficient mouse.
RESULTS
LC/MS/MS Analysis of Caspase-14 Interacting Molecules
Extracts prepared from 1) growth phase of keratinocytes, 2) differentiated phase of keratinocytes, and 3) corneocytes scraped from heels were reacted with revC14 or GST-procaspase-14 and bound proteins were identified by means of LC/MS/MS analysis. Each analysis identified 30–50 peptides. Candidate molecules were selected based on the following criteria, (a) unique in 2; (b) unique in 3; (c) common in 2 and 3; and (d) not keratins or related proteins. Peptides commonly found in procaspase-14 were ignored. Table 1 summarizes active caspase-14 interacting molecules selected according to the above criteria. We focused on cerebroside sulfate activator protein (prosaposin or saposin A) and trypsinogen-4 (mesotrypsinogen). Because prosaposin is processed by protease(s) that have not yet been identified, we considered that caspase-14 and trypsinogen-4 (mesotrypsin) might be involved.
TABLE 1.
Summary of LC/MC/MC analysis for caspase-14 interacting molecules Protein ID probability
| No | Description | KCa (differentiated) extract |
KC (growth) extract |
CC extract, act C14 | ||
|---|---|---|---|---|---|---|
| Pro-C14 | act C14 | Pro-C14 | act C14 | |||
| 1 | Caspase-14 precursor | 3.32E-12 | 2.84E-13 | 5.31E-12 | 2.44E-13 | 6.59E-04 |
| 2 | β Actin | NDb | 1.17E-12 | ND | ND | ND |
| 3 | Hypothetical protein | ND | 6.93E-11 | ND | ND | ND |
| 4 | Ribosomal protein S3a | ND | 6.17E-09 | ND | ND | ND |
| 5 | DEAD (Asp-Glu-Ala-Asp) box polypeptide | ND | 9.89E-09 | ND | ND | ND |
| 6 | Poly(A)-binding protein, cytoplasmic 4 | ND | 1.05E-08 | ND | ND | ND |
| 7 | Ribosomal protein S15 | ND | 6.45E-08 | ND | ND | ND |
| 8 | OCIA domain containing 2 isoform 2 | ND | 8.77E-07 | ND | ND | ND |
| 9 | S100 calcium binding protein A10 | ND | 9.03E-07 | ND | ND | ND |
| 10 | Zinc finger protein 185 (LIM domain) | ND | 3.47E-06 | ND | ND | ND |
| 11 | Cerebroside sulfate activator | 8.43E-10 | 3.71E-06 | ND | ND | 2.55E-05 |
| 12 | Annexin A2 isoform 2 | ND | 5.41E-06 | ND | ND | ND |
| 13 | Glycoprotein (transmembrane) nmb isoform | ND | 9.13E-06 | ND | ND | ND |
| 14 | Ribosomal protein S17 | ND | 1.16E-05 | ND | ND | ND |
| 15 | Plakophilin 3 | ND | 2.31E-05 | ND | ND | ND |
| 16 | Ribosomal protein S4, X-linked X isoform | ND | 3.95E-05 | ND | ND | ND |
| 17 | Isocitrate dehydrogenase (NADP+) | ND | 2.88E-04 | ND | ND | ND |
| 18 | Protein phosphatase 1, catalytic subunit, b | ND | 4.69E-04 | ND | ND | ND |
| 19 | PH0268 epidermal autoantigen 450K | ND | 8.05E-04 | ND | ND | ND |
| 20 | Trypsinogen IV b-form | ND | ND | ND | ND | 1.50E-04 |
a KC, keratinocyte; CC, cornified cell; Pro-C14, procaspase-14; act C14, active caspase-14.
b ND, not determined.
Confirmation of Prosaposin and Saposin A by Western Blotting and Immunohistochemistry
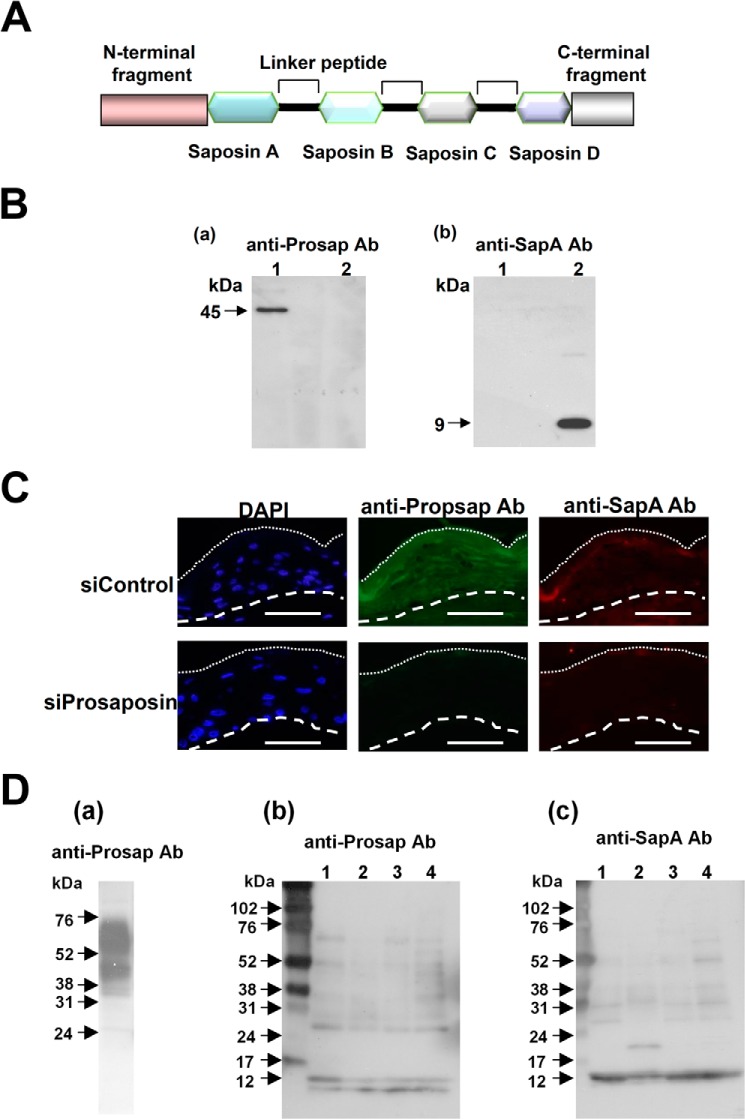
Schematic illustration of the prosaposin structure is shown in Fig. 1A. Because LC/MS/MS analysis identified a part of the saposin A sequence, EIVDSYLPVILDIIK, as a caspase-14 interactor, we raised an antibody to saposin A. Anti-SapA Ab was highly specific for saposin A (Fig. 1B). Also, anti-Prosap Ab did not recognize saposin A. Specificity of anti-Prosap Ab and anti-SapA Ab was further confirmed with immunohistochemcal analysis using skin equivalent models under the prosaposin knockdown conditions (Fig. 1C). Anti-Prosap Ab showed positive staining in the entire epidermis, whereas anti-Sap A demonstrated confined staining at the granular layer. Knockdown of prosaposin abolished these stainings, indicating highly specific features of these antibodies. If prosaposin is processed during terminal differentiation, we would expect degradation products and mature saposins to be detected in cornified cell extracts. Therefore, we investigated the presence of prosaposin and related products in extract from skin equivalent models and in cornified cell extracts (Fig. 1D). Heavy staining of prosaposin 60- and 45-kDa bands was detected in the skin equivalent extract, in addition to some smaller bands. The 60-kDa band is considered to be intact prosaposin with glycosyl side chains. In contrast, cornified cell extracts did not contain this band, but showed many smaller bands recognized by anti-Prosap Ab. The differences may indicate that processing of prosaposin is not uniform, but varies from individual to individual. Anti-SapA Ab also detected several bands that appeared to be degradation products. One of them, a 12-kDa band dominantly found in all specimens, appears to correspond to mature saposin A with carbohydrate side chains (13). These results are consistent with the idea that saposin A generation involves multiple steps and may be rather variable depending upon the environment.
FIGURE 1.
Detection of prosaposin and saposin A. A, a schematic illustration of prosaposin structure is shown. B, reactivity of anti-Prosap and anti-SapA antibodies (Ab). Specificities of these antibodies were tested with recombinant prosaposin (lane 1) and saposin A (lane 2). Western blot analysis was carried out using antibodies to prosaposin (anti-Prosap Ab) and saposin A (anti-SapA Ab). Anti-Prosap Ab recognized a 45-kDa band that is a non-glycosylated form of prosaposin. Anti-SapA Ab recognized saposin A at 9 kDa. C, immunohistochemical analysis of the antibody specificity. Skin equivalent models were constructed after treatment with nonspecific siRNA control (siControl) and specific siRNA to prosaposin (siProsaposin). Sections were stained with anti-Prosap Ab or anti-SapA Ab. Nuclear staining with DAPI was also shown. Dotted line shows the border of the cornified layer. Broken line indicates the epidermal-dermal junction. Scale bars: 50 μm. D, presence of prosaposin and saposin A in cornified cell extracts. a, extracts from a skin equivalent model were used as a prosaposin control. Heavy staining of the 65- (glycosylated form) and 45-kDa bands were evident with some degradation products. b, extracts from cornified cells of healthy subjects (lanes 1–4) were subjected to Western blotting using anti-Prosap Ab. Anti-Prosap Ab recognized the liberated N-terminal fragments with 10 and 12 kDa as well as intermediate degradation products. c, the presence of saposin A in the same extracts was tested with anti-SapA Ab. Each lane contained 10 μg of extracts.
Prosaposin Processing by Epidermal Proteases
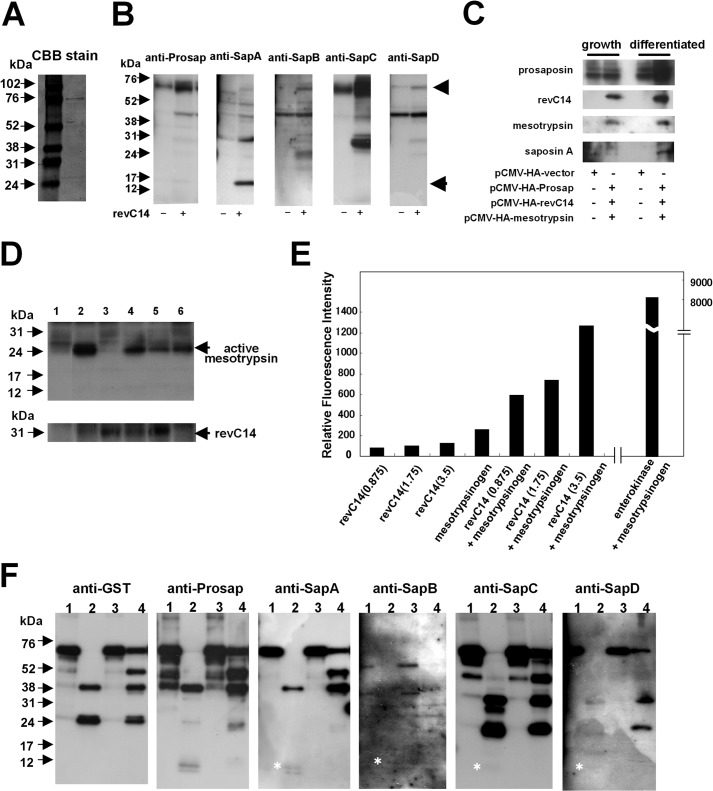
To examine the mechanism of saposin A generation, we examined epidermal proteases including caspase-14, mesotrypsin, KLK5, and KLK7. First we tested the purity of the recombinant GST-prosaposin (Fig. 2A). A band of ∼75 kDa was detected with Coomassie Brilliant Blue staining, indicating that the recombinant protein is suitable for use in analysis of prosaposin processing. To examine the effect of active caspase-14, we prepared constitutively active caspase-14 (revC14), which has equivalent activity to purified human caspase-14 (6). revC14 was incubated with extracts obtained from differentiated keratinocytes (Fig. 2B). We detected a band at 15 kDa that corresponds to the glycosylated form of saposin A. Some other bands apparently corresponding to intermediates were also detected. We further tested SapA generation after co-transfection of pCMV-HA-Prosap, pCMV-HA-revC14, and pCMV-HA-mesotrypsin in growth and differentiated phases (Fig. 2C). In a growing condition, co-expression of these three molecules did not yield detectable SapA. In contrast, co-expression results in the generation of SapA in the differentiated condition. Because we had verified caspase-14 as an interactor with saposin and mesotrypsin in the LC/MS/MS analysis, we investigated whether caspase-14 shows any effect on mesotrypsin in cultured keratinocytes. When revC14 was expressed in differentiated keratinocytes, the active form of mesotrypsin was strongly up-regulated (Fig. 2D, lane 2). The active form was not detected in the presence of pan-caspase inhibitor, Z-VAD-fmk (lane 3). A serine protease inhibitor, leupeptin, did not suppress generation of the active form of mesotrypsin, although it had a minor effect (lane 4). These results indicate that caspase-14 may function in mesotrypsinogen activation. Therefore we tested the activator activity of caspase-14 on mesotrypsinogen and compared it with the action of enterokinase, a well known trypsin activator. Incubation of mesotrypsinogen with revC14 resulted in a linear increase of mesotrypsin activity, although its activity was only of that of enterokinase (Fig. 2E). Thus, caspase-14 may contribute to the conversion of mesotrypsinogen to active mesotrypsin.
FIGURE 2.
Effect of epidermal proteases on prosaposin processing. A, Coomassie Brilliant Blue staining of GST-prosaposin. Recombinant GST-prosaposin was subjected to SDS-PAGE and stained with Coomassie Brilliant Blue. The band of the recombinant protein can be seen at ∼75 kDa. B, Western blot analysis of prosaposin degradation products. Extracts from differentiated keratinocytes containing prosaposin were incubated with revC14. Western blot analysis were carried out using antibodies to prosaposin (anti-Prosap), saposin A (anti-SapA), sapoins B (anti-SapB), saposin C (anti-SapC), and saposin D (anti-SapD). During incubation with active caspase-14, prosaposin in the extract was degraded into multiple intermediate products. The glycosylated form of saposin A (15 kDa) was detected. Arrowhead, GST-prosaposin; arrow, saposin A. C, co-transfection of pCMV-HA-Prosap, pCMV-HA-revC14, and pCMV-HA-mesotrypsin in growth and differentiated phases. The Western blot was carried out using a specific antibody to each molecule. D, detection of the active form of mesotrypsin in revC14-transfected keratinocytes. Keratinocytes were transfected with pCMV-HA-vector (control), pCMV-HA-revC14, or pCMV-HA-mesotrypsin and further incubated for 24 h in the presence or absence of protease inhibitors. Cell extracts were subjected to SDS-polyacrylamide gel electrophoresis and the presence of mesotrypsin was analyzed by Western blotting using anti-mesotrypsin antibody. Lane 1, pCMV-HA-vector; lane 2, pCMV-HA-revC14; lane 3, pCMV-HA-revC14 + Z-VAD-fmk; lane 4, pCMV-HA-revC14 + leupeptin; lane 5, pCMV-HA-revC14 + Z-VAD-fmk + leupeptin; lane 6, pCMV-HA-mesotrypsin. E, effect of caspase-14 on mesotrypsinogen activation. Enzymatic activity of mesotrypsin was measured using Boc-Gln-Ala-Arg-methylcoumarin amide as a substrate after incubation with caspase-14. To evaluate the direct hydrolytic activity of caspase-14 on this substrate, the same concentration of caspase-14 was incubated without mesotrypsinogen. Amounts of enzymes used in each assay (ng) are listed in parentheses. For comparison, enterokinase was also used. Results are shown as the mean of duplicate experiments. F, Western blot analysis of prosaposin degradation products by mesotrypsin, KLK5, and KLK7. After incubation with each protease, prosaposin degradation products were detected with antibodies to GST, prosaposin, saposin A, saposin B, saposin C, and saposin D. Asterisks indicate the presence of each saposin protein band. Lane 1, prosaposin control; lane 2, prosaposin + mesotrypsin; lane 3, prosaposin + KLK5; lane 4, prosaposin + KLK7.
These results also suggest that prosaposin processing could be accomplished by mesotrypsin. Indeed, when mesotrypsin was incubated with GST-prosaposin, various fragments were generated and detected with anti-saposin antibodies, in addition to anti-GST Ab (Fig. 2F). The reactivity with anti-SapA Ab indicated that saposin A-related products are formed by mesotrypsin digestion, and saposin A itself (12 kDa) was liberated by mesotrypsin (Fig. 2F, anti-SapA, lane 2). Furthermore, we could detect formation of all the other saposins after incubation with mesotrypsin, although the bands were considerably weaker than that of saposin A. Putative intermediate fragments of higher molecular weight were also detected with each saposin antibody. These results again seem consistent with a multistep process of saposin generation. Interestingly, KLK7 generated several intermediate products from prosaposin; some were similar to those formed by mesotrypsin and others were unique to KLK7. In contrast, KLK5 did not generate any saposin-related products from prosaposin.
Expression and Localization of Prosaposin and Saposin A
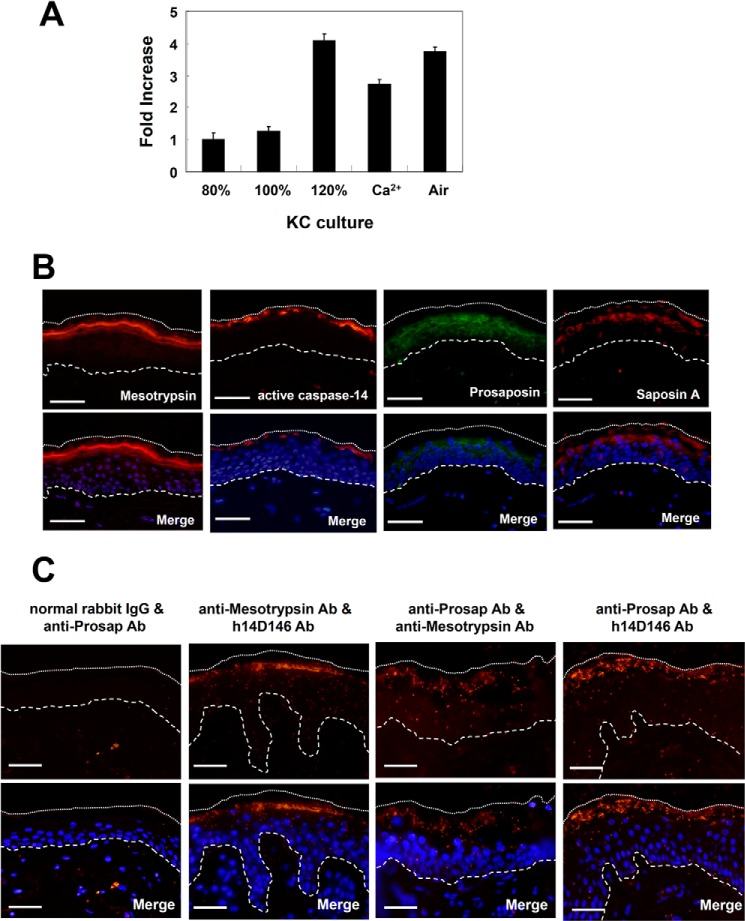
Expression of prosaposin in cultured keratinocytes was examined by means of quantitative PCR. Synthesis of prosaposin mRNA was quite low in growing keratinocytes, but was up-regulated in differentiated keratinocytes, e.g. during prolonged culture after confluence, in the presence of calcium and after air exposure (Fig. 3A). Prosaposin was localized in the region from basal to granular cells with slightly more intense staining in the granular layer (Fig. 3B). Staining with anti-SapA Ab was confined to granular cells. The mature form of caspase-14 was found in some granular cells and stratum corneum (SC). Mesotrypsin(ogen) was detected strongly in stratum granulosum (SG). These findings are in accordance with previous reports (6, 7). The presence of these proteases and saposin A at the granular layer indicates that this may be the site of saposin A generation. Using the antibody combinations of anti-mesotrypsin Ab/h14D146, anti-Prosap Ab/anti-mesotrypsin Ab, and anti-Prosap Ab/h14D146, PLA was carried out to investigate the in vivo interaction of these molecules in normal human skin (Fig. 3C). We used h14D146, because this antibody specifically recognizes the active form of caspase-14 (4). Specificity of this method is verified using normal rabbit IgG and anti-Prosap IgG. The antibody combination of mesotrypsin and active caspase-14 showed confined interaction at the SG. Prosaposin and mesotrypsin indicated positive reactions from the lower to upper epidermis, with a punctuate pattern especially at the SG. The antibody combination of prosaposin and active caspase-14 demonstrated strong interaction at the SG and SC. These results clearly showed that prosaposin and these proteases are associated in the SG to SC regions of normal human epidermis.
FIGURE 3.
Expression and localization of prosaposin and saposin A. A, expression of prosaposin gene transcript in cultured keratinocytes. cDNAs were prepared from cultured keratinocytes at 80% confluence, 100% confluence, 2 days after confluence (120%), 2 days after confluence in the presence of 1.2 mm CaCl2, and 2 days after confluence together with air exposure. The prosaposin mRNA levels were determined using real-time RT-PCR. Data were normalized to the GAPDH gene. B, immunohistochemical localization of mesotrypsin, active caspase-14, prosaposin, and saposin A. Nuclei were counterstained with DAPI. Merged figures of antibody staining images with nuclear staining images are also shown. The dotted line shows the edge of the cornified layer. The broken line shows the epidermal-dermal junction. Scale bars: 50 μm. C, demonstration of interaction by PLA. Interaction between prosaposin, active caspase-14, and mesotypsin was investigated in vivo using the PLA method. Detection was carried out using the following antibody combinations: normal rabbit IgG/anti-Prosap Ab (negative control), anti-mesotrypsin Ab/h14D146, anti-Prosap Ab/anti-mesotrypsin Ab, and anti-Prosap Ab/h14D146. Merged figures of antibody staining images with nuclear staining images are also shown. Scale bars, 50 μm.
Effect of siRNAs Specific to Caspase-14 and Mesotrypsin
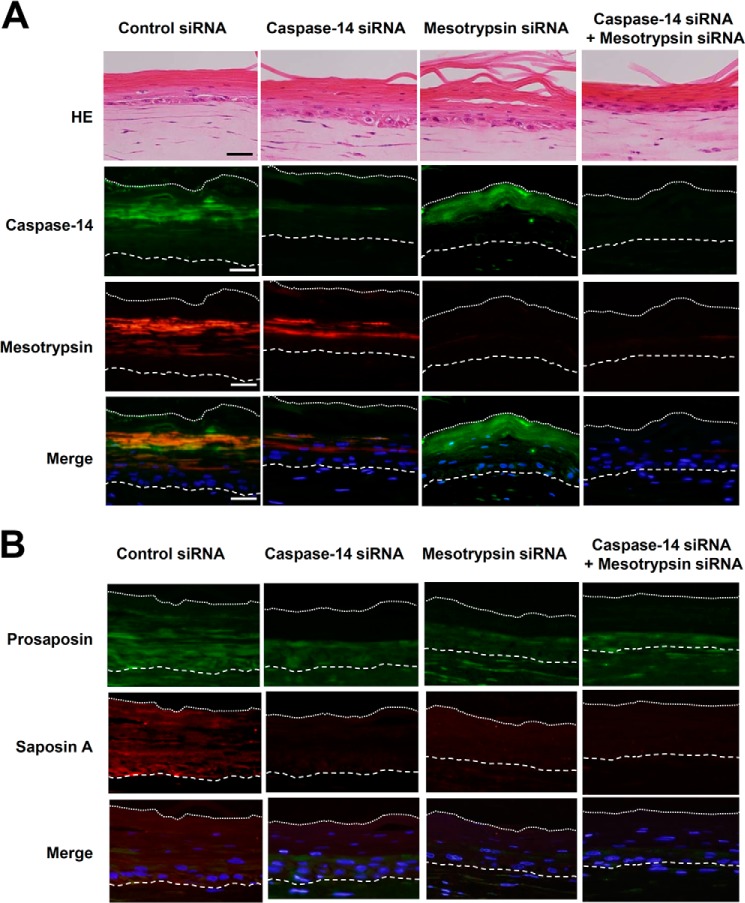
To further investigate the involvement of caspase-14 and mesotrypsin in saposin A generation, we constructed skin equivalent models with knockdown of caspase-14, mesotrypsin, or both. The effects of siRNAs on target proteases were examined with quantitative real-time PCR. Both siRNAs caused significant down-regulation of the expression of caspase-14 and mesotrypsin, to 9.9 and 24% of the control levels, respectively. When growing keratinocytes were treated with control siRNA and seeded on the dermal matrix, the resulting skin models showed considerable expression of caspase-14, mesotrypsin (Fig. 4A), and prosaposin and saposin A (Fig. 4B). All of these molecules were broadly distributed in the skin equivalent models. In particular, saposin A was not restricted to the boundary area, but was localized from the basal to cornified layers. Caspase-14 siRNA greatly reduced caspase-14 expression, but did not affect expression of mesotrypsin or prosaposin. Interestingly, saposin A was hardly detectable. Mesotrypsin siRNA showed similar effects, without influencing the expression of caspase-14 or prosaposin (except saposin A). Double knockdown of caspase-14 and mesotrypsin strongly down-regulated these proteases as well as saposin A, although prosaposin expression was not significantly changed. These results further support involvement of caspase-14 and mesotrypsin in generation of saposin A.
FIGURE 4.
Effects of knockdown of caspase-14 and mesotrypsin on skin equivalent models. A, immunohistochemical analysis of the expression of caspase-14 and mesotrypsin. After treatment of keratinocytes with nonspecific control siRNA, caspase-14 siRNA, mesotrypsin siRNA, or the combination of both, keratinocytes were seeded on dermal components and skin equivalent models were constructed. Thin sections were stained with caspase-14 mAb and anti-mesotrypsin Ab. HE, hematoxylin and eosin stain. B, down-regulation of saposin A in the skin equivalent models after knockdown of caspase-14 and mesotrypsin. Sections from skin equivalent models treated with appropriate siRNAs were stained with anti-Prosap Ab and anti-SapA Ab. Merged figures of antibody staining images with nuclear staining images are also shown. Scale bars, 50 μm.
Oil Red Staining of Skin with Prosaposin-deficient and Saposin A-deficient Mouse
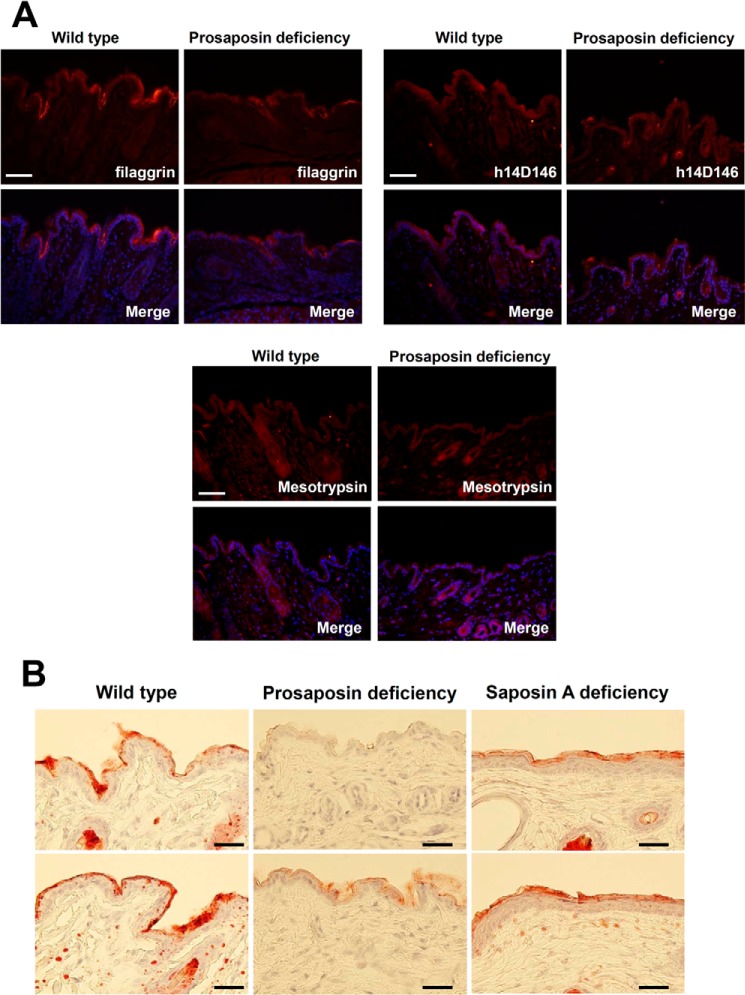
Next we examined how defects in prosaposin processing affect epidermal structure and permeability barrier formation. For this purpose, we examined skin from prosaposin-deficient and saposin A-deficient mice. First, we evaluated expression of filaggrin, active caspase-14, and mesotrypsin in prosaposin-deficient mice (Fig. 5A). Expression of filaggrin was slightly decreased. In contrast, expression of active caspase-14 or mesotrypsin remained unchanged. Next, we examined changes of lipid content (Fig. 5B). In normal mouse skin, Oil Red staining gave an intense positive reaction in the cornified layer, indicating the effectiveness of this method for staining the lipid envelope of cornified cells. In contrast, a positive reaction was hardly detectable in the epidermis of prosaposin-deficient mouse. Staining was significantly reduced in the cornified layer of saposin A-deficient mouse.
FIGURE 5.
Prosaposin and saposin A deficiency affects lipid content in the cornified layer. A, immunohistochemical analysis of filaggrin, active caspase-14, and mesotrypsin expression in wild-type and prosaposin-deficient mice. Merged figures of antibody staining images with nuclear staining images are also shown. Scale bars, 50 μm. B, Oil Red staining of skin of prosaposin-deficient and saposin A-deficient mice. Two skin examples each are shown for wild-type mice, prosaposin-deficient mice, and saposin A-deficient mice. Scale bars, 50 μm.
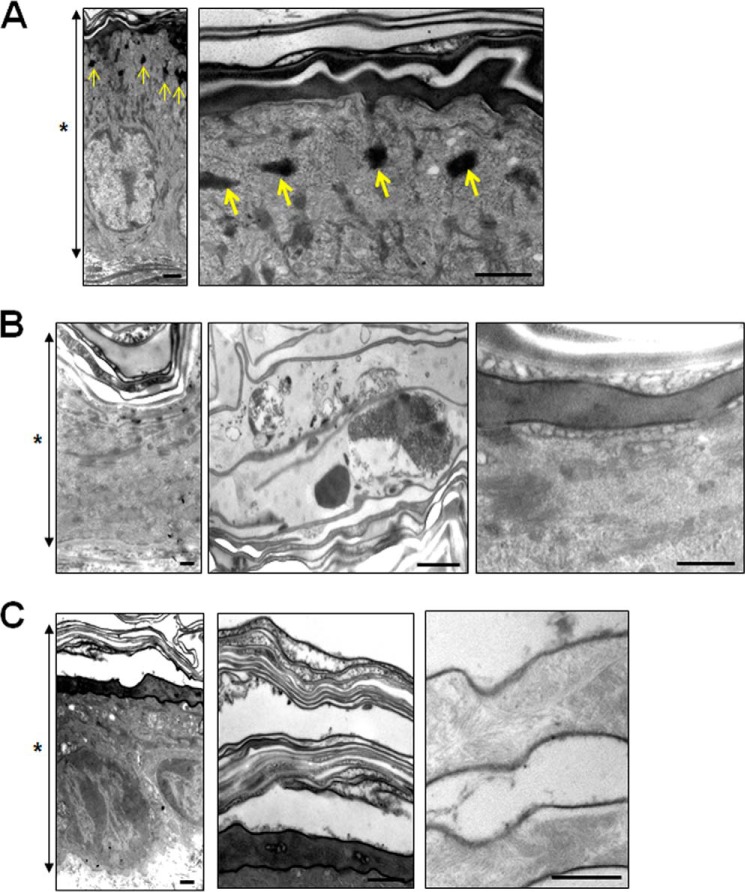
Ultrastructural Observation
Finally, we observed the epidermal ultrastructure of prosaposin-deficient and saposin A-deficient mice. Keratohyaline granules were clearly observed in wild-type mouse epidermis (Fig. 6A). In contrast, keratohyaline granules were rarely found in prosaposin-deficient mouse epidermis. Secreted lamellar granules surrounded the granular cells and retained a spherical pattern. Cell-cell contact between corneocytes was lost. In corneocytes, no keratin pattern remained and cell debris occupied the intercellular spaces (Fig. 6B). In a saposin A-deficient mouse, the epidermal structure seemed slightly more intact than in the prosaposin-deficient mouse. However, keratohyaline granules were hardly detectable and there was no clear keratin pattern in the corneocytes (Fig. 6C).
FIGURE 6.
Ultrastructural observation of skin from prosaposin-deficient and saposin A-deficient mouse. Skin from a wild-type mouse (A), prosaposin-deficient mouse (B), and saposin A-deficient mouse (C) was examined by electron microscopy. A, keratohyaline granules (arrows) were clearly observed in the wild-type mouse epidermis. Note that keratohyaline granules were rarely found in prosaposin-deficient and saposin A-deficient mice in the low magnification views (B and C). B, high magnification views of the cornified layer and transitional area from granular to cornified cell layers in the epidermis of prosaposin-deficient mouse are also shown. C, low magnification views of the skin (left) and entire cornified layer (middle), and higher magnification of cornified cells in the lower layer of saposin A-deficient mouse (right) are shown. Asterisks show the entire epidermis. Scale bars, 100 nm.
DISCUSSION
We employed LC/MS/MS analysis to identify molecules interacting with caspase-14. This technique has extraordinary sensitivity, and to exclude nonspecific interactors and indirect interactors, we compared sets of affinity purified samples obtained from extracts of keratinocytes at different phases as well as cornified cell extracts with GST fusion active caspase-14 or procaspase-14. We focused on prosaposin and mesotrypsin(ogen) (PRSS3), because they were detected as caspase-14 interactors and were found only in extracts from differentiated keratinocytes and cornified cells. We hypothesized that caspase-14 and mesotrypsin are involved in prosaposin processing.
However, caspase-14, either in constitutively active form (revC14) (4) or intermediate form (revC14-Y178) (6), did not show any direct effect on saposin A generation. Instead, unexpectedly, we found that caspase-14 has the ability to convert mesotrypsinogen to mesotrypsin. This is probably accomplished via cleavage near the recognition sequence for enterokinase (DDDDKI) (23), because caspase-14 cleaves only after Asp residues. To our knowledge, this is the first observation that caspase-14 works as an activator of a protease cascade. The caspase-14 concentration used in this study was comparable with that of enterokinase. The catalytic efficiency of enterokinase is 34,000-fold greater than that of trypsin (24). Thus, even though caspase-14 was severalfold less active than enterokinase as an activator, this would not exclude a role of caspase-14 as a possible activator in vivo. However, it is not yet clear whether caspase-14 participates in mesotrypsinogen activation in vivo. In our previous study, we showed that enterokinase is expressed at the boundary area. Because mesotrypsin(ogen) is, at least in part, associated with caspase-14 as shown by LC/MS/MS analysis, and caspase-14 requires a high concentration of kosmotropic ions for its activation, enterokinase and caspase-14 may share a role, with one or the other being predominant, depending on the environment.
As far as saposin processing is concerned, it is suggested that generation of four saposins from prosaposin occurs by cleavage at Lys or Arg (13), indicating involvement of trypsin-like protease(s). Mesotrypsin would be a candidate for the maturation of saposins, including saposin A. Epidermal mesotrypsin was cloned from a keratinocyte cDNA library as a unique splicing variant (7). Expression and localization of enterokinase were confirmed at the granular layer of human epidermis, and an enterokinase cleavage fragment was co-localized at the same site (7). Interestingly, mesotrypsin is not susceptible to intrinsic trypsin inhibitors and exhibits low degradation activity toward high molecular weight proteins (25). These unique features of mesotrypsin may be consistent with a role in limited proteolysis, including protein processing.
We also tested the effect of trypsin-like serine protease KLK5, but no saposin A-related band was produced when prosaposin was incubated with KLK5 alone. Instead, KLK7 caused limited proteolysis of prosaposin, generating some intermediate bands. It is not yet clear whether KLK7 is also involved in this reaction. However, the fact that KLK7 is able to activate procaspase-14 (6) suggests that it may have a role in prosaposin processing.
Involvement of caspase-14 and mesotrypsin in prosaposin processing was further confirmed by the use of skin equivalent models with knockdown of these proteases. Although caspase-14, mesotrypsin, prosaposin, and saposin A showed broader distributions compared with normal human skin, these proteins were also present in epidermal components. Interestingly, knockdown of either caspase-14 or mesotrypsin, as well as double knockdown of these proteases, greatly diminished saposin A expression (Fig. 4B). These results suggest that caspase-14 and mesotrypsin work cooperatively for the production of saposin A. Indeed, a close association between mature caspase-14, mesotrypsin, and prosaposin was also demonstrated in normal human skin by the PLA method.
Functional importance of prosaposin processing was demonstrated by studying prosaposin-deficient and saposin A-deficient mice. Oil Red staining showed that loss of prosaposin significantly reduced the lipid envelope formation in skin of prosaposin- and saposin A-deficient mice. These results suggest that impairment of prosaposin processing leads to a defect of lipid envelope formation, consequently disrupting permeability barrier function. Electron microscopic observation confirmed key roles of prosaposin and saposin A in maintaining epidermal structure. Doering et al. (26) reported the impaired ultrastructure of skin in a newborn prosaposin-deficient mouse (27). We used skin from a 1-month-old deficient mouse and observed a profound effect of prosaposin deficiency. Not only cornified cells but also granular cells exhibited abnormal structure. Keratohyaline granules were almost absent in the skin of the prosaposin-deficient mouse, and similar results were obtained for a saposin A-deficient mouse. It is not yet clear why loss of saposins affects keratohyaline granule formation, but this may further impair epidermal barrier function.
Earlier reports indicated that prosaposin processing is dependent on lysosomal hydrolysis (27). It is not known whether prosaposin processing occurs in lysosomes before secretion, or in intercellular spaces in human epidermis. Whichever is the case, a previous immunoelectron microscopy study clearly showed that caspase-14 was localized not only in cytoplasm, but also in intercellular spaces in the granular to cornified layers (28). Our study also revealed that epidermal mesotrypsin is similarly present both inside and outside the granular to cornified cell layers (29). Thus, we postulate that caspase-14 and mesotrypsin both participate in processing of prosaposin. The specific antibody to saposin A prepared in this study revealed saposin A staining at the transition phase from granular to cornified cells. This is the site of extensive hydrolytic degradation and re-organization of cellular components to form corneocytes that function as a part of the protective barrier. On the other hand, many kinds of enzymes, such as desquamation proteases (KLKs) (30), natural moisturizing factor-generating enzymes (31), and filaggrin processing proteases (32) are synthesized at this stage. Synthesis and activation of caspase-14 and mesotrypsin also occur in the same area (4, 7). Because keratinocyte terminal differentiation is a unique process involving specialized molecules that are not found in other tissues, processing of prosaposin may also be regulated by differentiation-specific proteases.
It has been established that deficiency or dysfunction of prosaposin or saposins can result in lysosomal storage disorders that can be mimicked by deficiency of the enzyme activated by the particular saposin(s) involved (14, 17). Total deficiency of human or mouse prosaposin results in aberrant storage of multiple glycosphingolipids in a variety of organs (33). Recent observations showed that saposin A deficiency leads to progressive encephalopathy and abnormal myelination in the cerebral white matter, as seen in Krabbe disease (22). Saposin C has particular relevance to Gaucher disease (34). On the other hand, specific deficiency of saposin D has not been reported in humans. It would be interesting to know whether tissue-specific mesotrypsin is involved in similar physiological reactions, because different levels of mesotrypsin expression are observed in different tissues (7).
In conclusion, we have uncovered a novel function of caspase-14 as an activator of mesotrypsinogen, and we have shown that epidermal mesotrypsin serves as a processing enzyme for prosaposin. Down-regulation of these proteases and dysregulation of prosaposin activation would result in impaired permeability barrier function. We believe these findings contribute to our understanding of keratinocyte biology, especially in relationship to diseases associated with aberrant barrier function, including atopic dermatitis and psoriasis.
Footnotes
- KLK
- kallikrein-related peptidase
- PLA
- proximity ligation assay
- SC
- stratum corneum
- SG
- stratum granulosum
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone.
REFERENCES
- 1. Van de Craen M., Van Loo G., Pype S., Van Criekinge W., Van den brande I., Molemans F., Fiers W., Declercq W., Vandenabeele P. (1998) Identification of a new caspase homologue: caspase-14. Cell Death Differ. 5, 838–846 [DOI] [PubMed] [Google Scholar]
- 2. Eckhart L., Declercq W., Ban J., Rendl M., Lengauer B., Mayer C., Lippens S., Vandenabeele P., Tschachler E. (2000) Terminal differentiation of human keratinocytes and stratum corneum formation is associated with caspase-14 activation. J. Invest. Dermatol. 115, 1148–1151 [DOI] [PubMed] [Google Scholar]
- 3. Denecker G., Hoste E., Gilbert B., Hochepied T., Ovaere P., Lippens S., Van den Broecke C., Van Damme P., D'Herde K., Hachem J. P., Borgonie G., Presland R. B., Schoonjans L., Libert C., Vandekerckhove J., Gevaert K., Vandenabeele P., Declercq W. (2007) Caspase-14 protects against epidermal UVB photodamage and water loss. Nat. Cell. Biol. 9, 666–674 [DOI] [PubMed] [Google Scholar]
- 4. Hibino T., Fujita E., Tsuji Y., Nakanishi J., Iwaki H., Katagiri C., Momoi T. (2010) Purification and characterization of active caspase-14 from human epidermis and development of the cleavage site-directed antibody. J. Cell Biochem. 109, 487–497 [DOI] [PubMed] [Google Scholar]
- 5. Lippens S., Kockx M., Knaapen M., Mortier L., Polakowska R., Verheyen A., Garmyn M., Zwijsen A., Formstecher P., Huylebroeck D., Vandenabeele P., Declercq W. (2000) Epidermal differentiation does not involve the pro-apoptotic executioner caspases, but is associated with caspase-14 induction and processing. Cell Death Differ. 7, 1218–1224 [DOI] [PubMed] [Google Scholar]
- 6. Yamamoto M., Miyai M., Matsumoto Y., Tsuboi R., Hibino T. (2012) Kallikrein-related peptidase-7 regulates caspase-14 maturation during keratinocyte terminal differentiation by generating an intermediate form. J. Biol. Chem. 287, 32825–32834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakanishi J., Yamamoto M., Koyama J., Sato J., Hibino T. (2010) Keratinocytes synthesize enteropeptidase and multiple forms of trypsinogen during terminal differentiation. J. Invest. Dermatol. 130, 944–952 [DOI] [PubMed] [Google Scholar]
- 8. Rawlings A. V., Harding C. R. (2004) Moisturization and skin barrier function. Dermatol. Ther. 17, 43–48 [DOI] [PubMed] [Google Scholar]
- 9. Proksch E., Holleran W. M., Menon G. K., Elias P. M., Feingold K. R. (1993) Barrier function regulates epidermal lipid and DNA synthesis. Br. J. Dermatol. 128, 473–482 [DOI] [PubMed] [Google Scholar]
- 10. Ishida-Yamamoto A., Iizuka H. (1998) Structural organization of cornified cell envelopes and alterations in inherited skin disorders. Exp. Dermatol. 7, 1–10 [DOI] [PubMed] [Google Scholar]
- 11. Dale B. A., Lonsdale-Eccles J. D., Holbrook K. A. (1980) Stratum corneum basic protein: an interfilamentous matrix protein of epidermal keratin. Curr. Probl. Dermatol. 10, 311–325 [DOI] [PubMed] [Google Scholar]
- 12. Jensen J. M., Proksch E. (2009) The skin's barrier. G. Ital. Dermatol. Venereol. 144, 689–700 [PubMed] [Google Scholar]
- 13. O'Brien J. S., Kishimoto Y. (1991) Saposin proteins: structure, function, and role in human lysosomal storage disorders. FASEB J. 5, 301–308 [DOI] [PubMed] [Google Scholar]
- 14. Kishimoto Y., Hiraiwa M., O'Brien J. S. (1992) Saposins: structure, function, distribution, and molecular genetics. J. Lipid Res. 33, 1255–1267 [PubMed] [Google Scholar]
- 15. Vaccaro A. M., Salvioli R., Tatti M., Ciaffoni F. (1999) Saposins and their interaction with lipids. Neurochem. Res. 24, 307–314 [DOI] [PubMed] [Google Scholar]
- 16. Grabowski G. A., Horowitz M. (1997) Gaucher's disease: molecular, genetic and enzymological aspects. Baillieres Clin. Haematol. 10, 635–656 [DOI] [PubMed] [Google Scholar]
- 17. Matsuda J., Yoneshige A., Suzuki K. (2007) The function of sphingolipids in the nervous system: lessons learnt from mouse models of specific sphingolipid activator protein deficiencies. J. Neurochem. 103, 32–38 [DOI] [PubMed] [Google Scholar]
- 18. Brattsand M., Stefansson K., Lundh C., Haasum Y., Egelrud T. (2005) A proteolytic cascade of kallikreins in the stratum corneum. J. Invest. Dermatol. 124, 198–203 [DOI] [PubMed] [Google Scholar]
- 19. Motoyama A., Venable J. D., Ruse C. I., Yates J. R., 3rd (2006) Automated ultra-high-pressure multidimensional protein identification technology (UHP-MudPIT) for improved peptide identification of proteomic samples. Anal. Chem. 78, 5109–5118 [DOI] [PubMed] [Google Scholar]
- 20. Motoyama A., Xu T., Ruse C. I., Wohlschlegel J. A., Yates J. R., 3rd (2007) Anion and cation mixed-bed ion exchange for enhanced multidimensional separations of peptides and phosphopeptides. Anal. Chem. 79, 3623–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fujita N., Suzuki K., Vanier M. T., Popko B., Maeda N., Klein A., Henseler M., Sandhoff K., Nakayasu H., Suzuki K. (1996) Targeted disruption of the mouse sphingolipid activator protein gene: a complex phenotype, including severe leukodystrophy and wide-spread storage of multiple sphingolipids. Hum. Mol. Genet. 5, 711–725 [DOI] [PubMed] [Google Scholar]
- 22. Matsuda J., Vanier M. T., Saito Y., Tohyama J., Suzuki K., Suzuki K. (2001) A mutation in the saposin A domain of the sphingolipid activator protein (prosaposin) gene results in a late-onset, chronic form of globoid cell leukodystrophy in the mouse. Hum. Mol. Genet. 10, 1191–1199 [DOI] [PubMed] [Google Scholar]
- 23. Light A., Janska H. (1989) Enterokinase (enteropeptidase): comparative aspects. Trends Biochem. Sci. 14, 110–112 [DOI] [PubMed] [Google Scholar]
- 24. Anderson R. G., Hein C. E. (1977) Distribution of anionic sites on the oviduct ciliary membrane. J. Cell Biol. 72, 482–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sahin-Tóth M. (2005) Human mesotrypsin defies natural trypsin inhibitors: from passive resistance to active destruction. Protein Pept. Lett. 12, 457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Doering T., Holleran W. M., Potratz A., Vielhaber G., Elias P. M., Suzuki K., Sandhoff K. (1999) Sphingolipid activator proteins are required for epidermal permeability barrier formation. J. Biol. Chem. 274, 11038–11045 [DOI] [PubMed] [Google Scholar]
- 27. Hiraiwa M., Martin B. M., Kishimoto Y., Conner G. E., Tsuji S., O'Brien J. S. (1997) Lysosomal proteolysis of prosaposin, the precursor of saposins (sphingolipid activator proteins): its mechanism and inhibition by ganglioside. Arch. Biochem. Biophys 341, 17–24 [DOI] [PubMed] [Google Scholar]
- 28. Alibardi L., Dockal M., Reinisch C., Tschachler E., Eckhart L. (2004) Ultrastructural localization of caspase-14 in human epidermis. J. Histochem. Cytochem. 52, 1561–1574 [DOI] [PubMed] [Google Scholar]
- 29. Miyai M., Matsumoto Y., Yamanishi H., Yamamoto-Tanaka M., Tsuboi R., Hibino T. (2014) Keratinocyte-specific mesotrypsin contributes to desquamation process via kallikrein activation and LEKTI degradation. J. Invest. Dermatol. 134, 1665–1674 [DOI] [PubMed] [Google Scholar]
- 30. Ovaere P., Lippens S., Vandenabeele P., Declercq W. (2009) The emerging roles of serine protease cascades in the epidermis. Trends Biochem. Sci. 34, 453–463 [DOI] [PubMed] [Google Scholar]
- 31. Kamata Y., Taniguchi A., Yamamoto M., Nomura J., Ishihara K., Takahara H., Hibino T., Takeda A. (2009) Neutral cysteine protease bleomycin hydrolase is essential for the breakdown of deiminated filaggrin into amino acids. J. Biol. Chem. 284, 12829–12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scharschmidt T. C., List K., Grice E. A., Szabo R. NISC Comparative Sequencing Program, Renaud G., Lee C. C., Wolfsberg T. G., Bugge T. H., Segre J. A. (2009) Matriptase-deficient mice exhibit ichthyotic skin with a selective shift in skin microbiota. J. Invest. Dermatol. 129, 2435–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vaccaro A. M., Motta M., Tatti M., Scarpa S., Masuelli L., Bhat M., Vanier M. T., Tylki-Szymanska A., Salvioli R. (2010) Saposin C mutations in Gaucher disease patients resulting in lysosomal lipid accumulation, saposin C deficiency, but normal prosaposin processing and sorting. Hum. Mol. Genet. 19, 2987–2997 [DOI] [PubMed] [Google Scholar]
- 34. Horowitz M., Zimran A. (1994) Mutations causing Gaucher disease. Hum. Mutat. 3, 1–11 [DOI] [PubMed] [Google Scholar]