FIGURE 4.
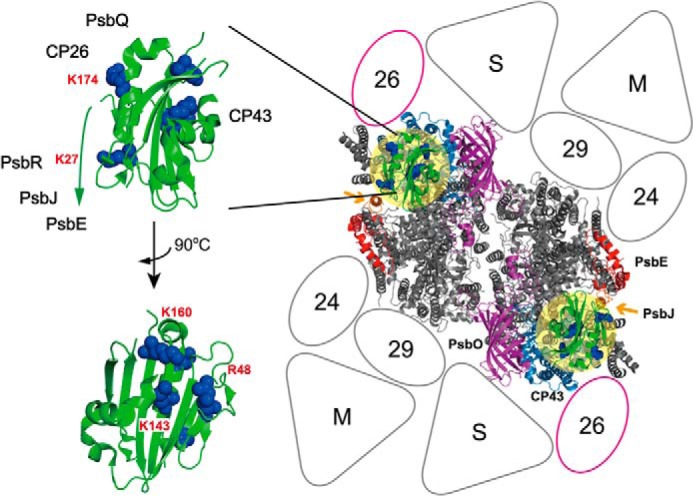
Proposed binding topology of PsbP in the LHCII-PSII supercomplex. Left, schematic of the PsbP structure from spinach (PDB ID 2VU4). The side chains of the residues Lys-27, Lys-K174, K160A, Lys-143, and Arg-48 involved in PsbP binding to PSII are shown as spherical models. Right, model of the LHCII-PSII supercomplex (C2S2M2 complex), viewed from the top onto the lumenal surface and utilizing the high resolution structure of the cyanobacterial PSII core (4), based upon comparisons with the literature (27, 30). Putative PsbP-binding sites are indicated by light yellow circular overlays. PsbV and PsbU, known to be absent in plant PSII, were excluded. CP43 and PsbE, which interact with PsbP, are shown in light blue and red, respectively. PsbJ and PsbO, required for the stable binding of PsbP and PsbQ, are shown in orange (indicated by arrows) and purple, respectively. Positioning of the strongly (S) or moderately (M) bound LHC trimers, as well as the minor LHC antennae CP24 (24), CP26 (26), and CP29 (29), are schematically depicted. CP26 is outlined in magenta to indicate its interaction with PsbP and PsbQ.
