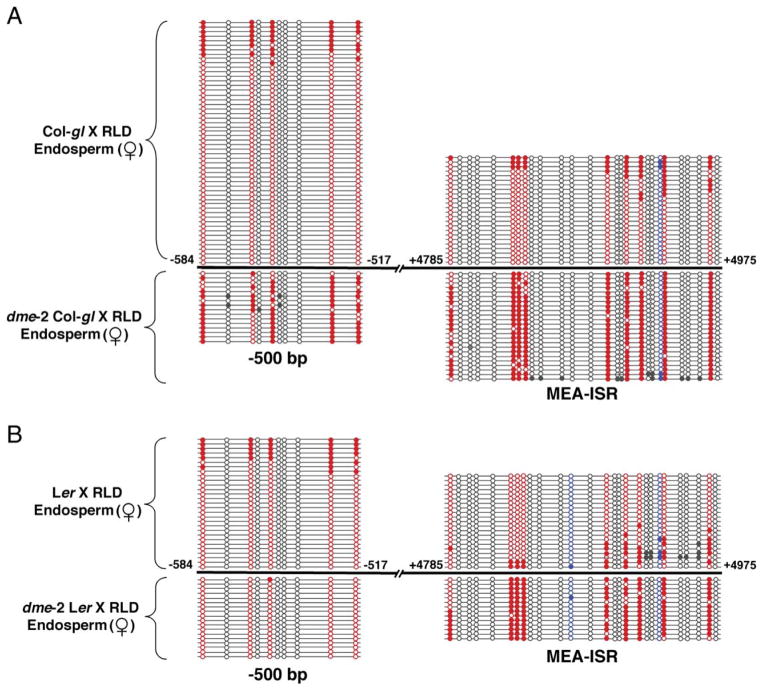
Figure 2. Hypermethylation of Maternal MEA in dme Mutant Endosperm.
Maternal-allele methylation in the −500 bp and MEA-ISR regions in endosperm from crosses between dme-2 heterozygous females and RLD males compared to maternal endosperm allele methylation from crosses between wild-type females and RLD males.
(A) dme-2 heterozygous Col-gl crossed to RLD.
(B) dme-2 heterozygous Ler crossed to RLD. Mutant endosperm was collected at 9 DAP from seeds with the dme endosperm overproliferation phenotype. Numbers are from the translation start site. To determine the pattern of DNA methylation, DNA was treated with bisulfite, PCR amplified, cloned, and sequenced. Circles connected by lines represent the results from determining the DNA sequence of one clone. Filled circle, methylated cytosine; open circle, unmethylated cytosine; red circle, CG site; blue circle, CNG site; gray circle, CNN site.