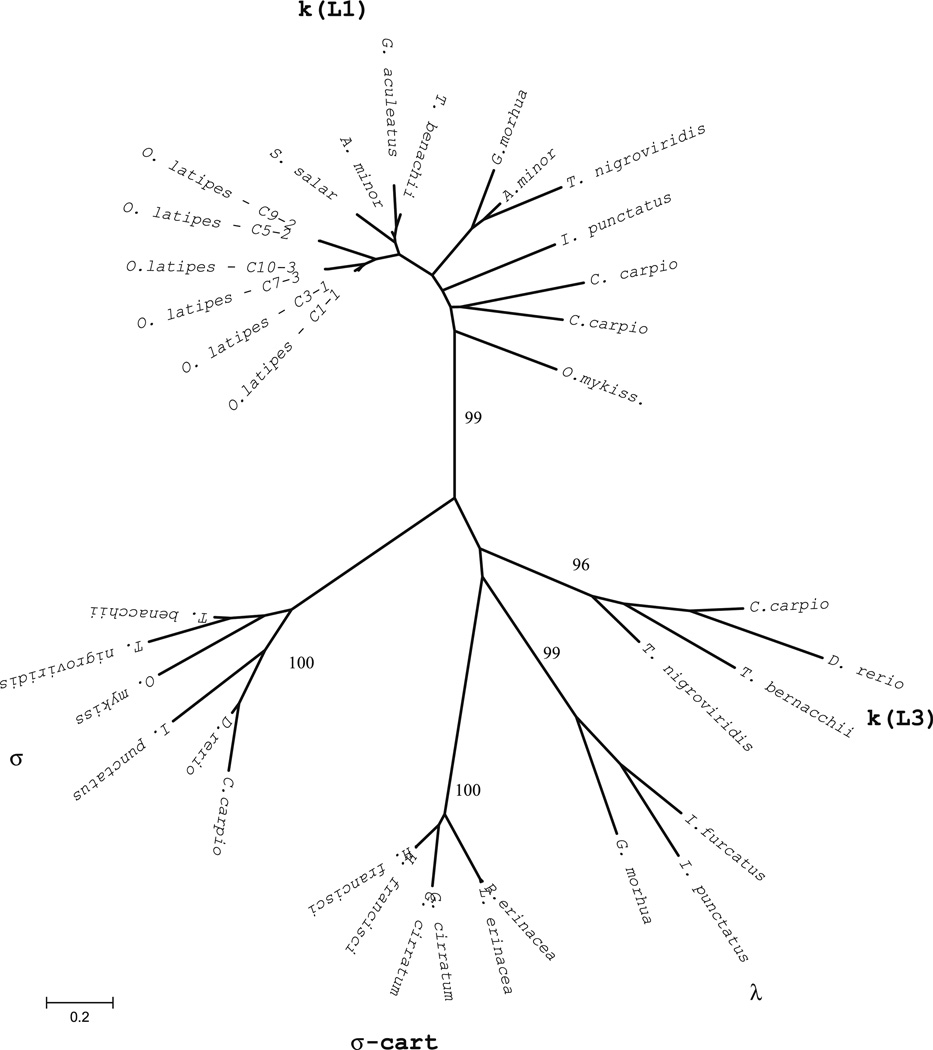
Fig. 3.
Phylogenetic analysis of medaka CL genes. The unrooted tree was constructed using two representative amino acid CL sequences from each medaka IGL locus and other identified fish CL sequences. The phylogenetic tree was constructed using the pairwise deletion algorithm, JTT matrix and differences by sites activated with gamma parameter 2.5. Sequences from other teleosts include GenBank accession numbers: kappa/Ll (G. aculeatus AY278356, T. bernacchii DQ842627, A. minor AF137397 and AF137398, S. salar AF406958, T. nigroviridis AJ575806, G morhua X68514, I. punctatus L25533, C. carpio AB015905 and X65260, O. mykiss AB035729), kappa/L3 (C. carpio AB035730, D. rerio AF246193, T. bernacchii DQ842626, T. nigroviridis U25705), lambda (I. furcatus CK403484; I. punctatus EU872022, G. morhua AJ293807), sigma (C. carpio AB091118, D. rerio AF246162, I. punctatus EU872021, T. nigroviridis AJ575637; T. bernacchii EF114785), sigma-cart (G. cirratum AAV34681 and AAV34682; N. Shark NS5; H. francisci XI5315 and B33937; L. erinacea L25568; E erinacea AAA87170)