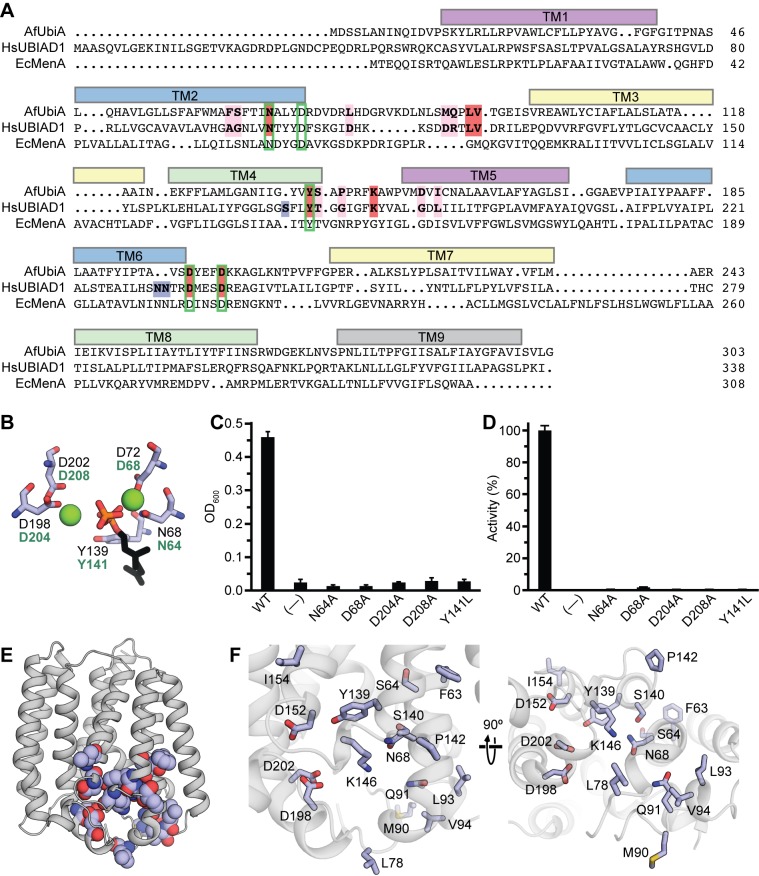
Figure 5. Conservation of active site residues across the UbiA family.
(A) A sequence alignment of AfUbiA, human UBIAD1, and EcMenA. Residues currently known to be mutated in SCD patients are highlighted in pink, red, or blue in the AfUbiA and HsUBIAD1 sequences. Red indicates mutated residues that are identical between the two proteins; blue indicates residues that align with gaps in the AfUbiA sequence. Green boxes indicate locations of residues that were mutated in the EcMenA functional assays. The colored bars above the alignment indicate locations of transmembrane helices in the AfUbiA crystal structure. (B) The locations of the active site residues marked with green boxes in panel (A) are shown relative to bound Mg2+ and GPP in the AfUbiA structure. Black labels correspond to residue numbers in the AfUbiA structure, green labels to the equivalent residues in EcMenA. (C) A menA − E. coli strain was transformed with plasmids containing either WT or mutant EcMenA, and grown in suspension cultures in an anaerobic chamber. The optical densities at 600 nm were measured after 24 h. Cells transformed with an unrelated protein (TrkH) were used as the negative control. Error bars are standard deviations of three experiments. Data used to calculate the bar graphs are shown in Table S2. (D) Membranes were purified from E. coli overexpressing WT or mutant EcMenA and incubated at 37°C for 10 min with 2 mM DHNA, 1 mM GPP, and 5 mM MgCl2. Product formation was measured by HPLC and is shown as a percentage of the activity for WT EcMenA. Membranes from cells overexpressing EcUbiA, which is selective for 4HB as the prenyl acceptor, were used as a negative control. Error bars are standard deviations of three experiments. Data used to calculate the bar graphs are shown in Table S3. (E) Residues currently known to be mutated in SCD patients are shown as spheres on the structure of AfUbiA. (F) The same residues are shown as sticks in a closer view of the substrate-binding cavity from within the plane of the membrane (left) and from the intracellular side (right).