Figure 5. Among residues encoded by the same codon within the same gene, those with higher demands for translational accuracy have lower elongation speeds. A total of 1,843 genes are used.
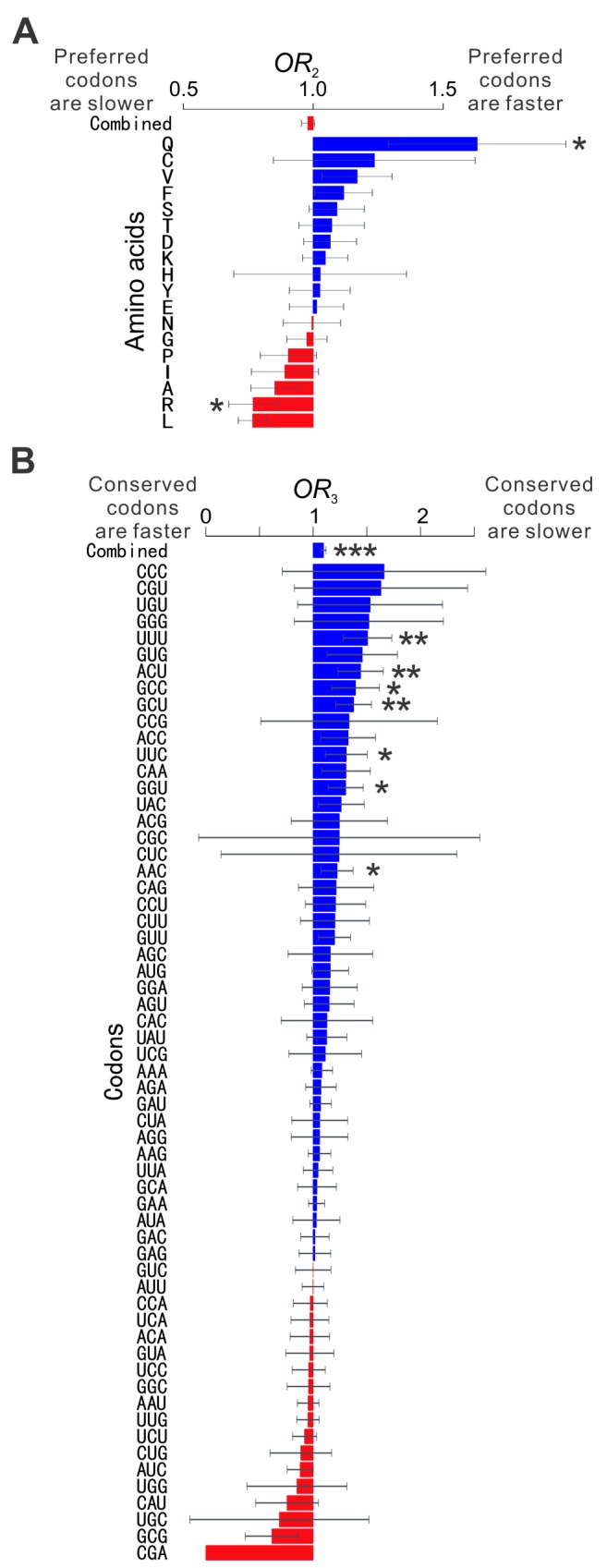
(A) Synonymous codon usage does not predict elongation speed. For each amino acid, OR 2 is calculated for each gene and then combined across genes by the MH procedure. OR 2>1 indicates that preferred codons are translated faster than unpreferred codons, and vice versa. The combined OR 2 from all amino acids is not significantly different from 1 (p>0.2). (B) Among residues encoded by the same codon in the same gene, those that are more conserved are translated more slowly. For each codon, OR 3 is calculated for each gene and then combined across genes by the MH procedure. OR 3>1 indicates that conserved residues encoded by a codon are translated more slowly than unconserved ones encoded by the same codon, and vice versa. The combined OR 3 from all codons is significantly greater than 1 (p<10−5). For both panels, error bar indicates one standard error, estimated by bootstrapping the genes 1,000 times. The standard error of OR 3 for CGA cannot be estimated because CGA with relevant information occurred in only one gene. Nominal p values from the MH test are indicated by asterisks. * p<0.05; ** p<0.01; *** p<0.001.
